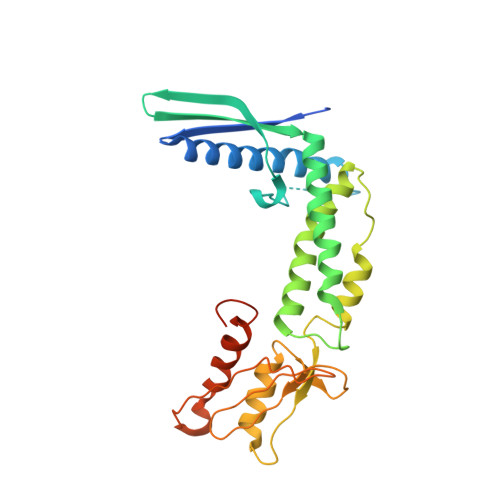
Crystal structure of the secreted protein HP1454 from the human pathogen Helicobacter pylori.
Quarantini, S., Cendron, L., Zanotti, G.(2014) Proteins 82: 2868-2873
- PubMed: 24854568
- DOI: https://doi.org/10.1002/prot.24608
- Primary Citation of Related Structures:
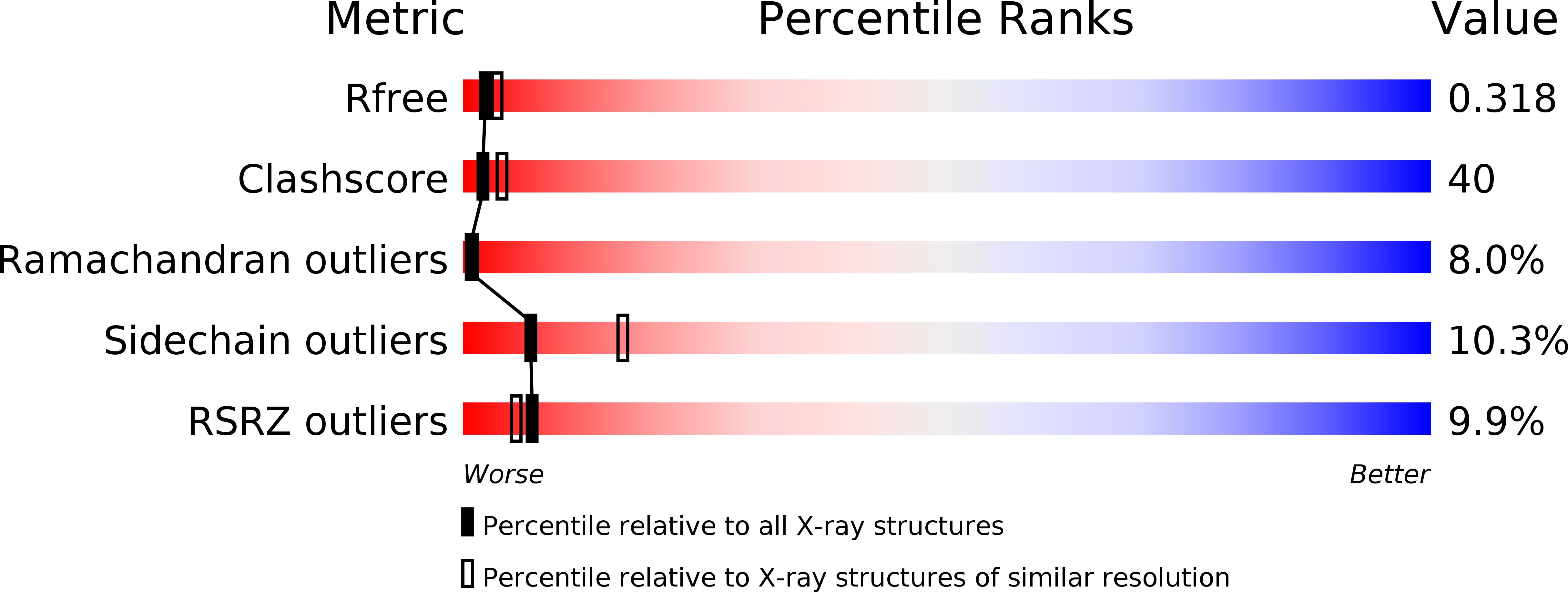
4KZS - PubMed Abstract:
HP1454 is a protein of 303 amino acids found in the extracellular milieu of Helicobacter pylori. The protein structure, crystallized in the orthorhombic C222₁ space group with one molecule per asymmetric unit, has been determined using the single-wavelength anomalous dispersion method. HP1454 exhibits an elongated bent shape, composed of three distinct domains. Each domain possesses a fold already present in other structures: Domain I contains a three-strand antiparallel β-barrel flanked by a long α-helix, Domain II is an anti-parallel three-helix bundle, and Domain III a β-sheet flanked by two α-helices. The overall assembly of the protein does not bear any similarity with known structures.
Organizational Affiliation:
Department of Biomedical Sciences, University of Padua, Viale G. Colombo 3, Padua, 35131, Italy.