Novel Modifications on C-terminal Domain of RNA Polymerase II Can Fine-tune the Phosphatase Activity of Ssu72.
Luo, Y., Yogesha, S.D., Cannon, J.R., Yan, W., Ellington, A.D., Brodbelt, J.S., Zhang, Y.(2013) ACS Chem Biol 8: 2042-2052
- PubMed: 23844594
- DOI: https://doi.org/10.1021/cb400229c
- Primary Citation of Related Structures:
4IMI, 4IMJ - PubMed Abstract:
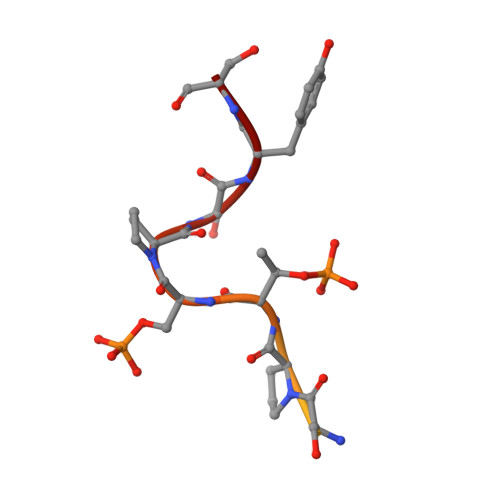
The C-terminal domain of RNA polymerase II (CTD) modulates the process of transcription through sequential phosphorylation/dephosphorylation of its heptide repeats, through which it recruits various transcription regulators. Ssu72 is the first characterized cis-specific CTD phosphatase that dephosphorylates Ser5 with a requirement for the adjacent Pro6 in a cis conformation. The recent discovery of Thr4 phosphorylation in the CTD calls into question whether such a modification can interfere with Ssu72 binding via the elimination of a conserved intramolecular hydrogen bond in the CTD that is potentially essential for recognition. To test if Thr4 phosphorylation will abolish Ser5 dephosphorylation by Ssu72, we determined the kinetic and structural properties of Drosophila Ssu72-symplekin in complex with the CTD peptide with consecutive phosphorylated Thr4 and Ser5. Our mass spectrometric and kinetic data established that Ssu72 does not dephosphorylate Thr4, but the existence of phosphoryl-Thr4 next to Ser5 reduces the activity of Ssu72 toward the CTD peptide by 4-fold. To our surprise, even though the intramolecular hydrogen bond is eliminated due to the phosphorylation of Thr4, the CTD adopts an almost identical conformation to be recognized by Ssu72 with Ser5 phosphorylated alone or both Thr4/Ser5 phosphorylated. Our results indicate that Thr4 phosphorylation will not abolish the essential Ssu72 activity, which is needed for cell survival. Instead, the phosphatase activity of Ssu72 is fine-tuned by Thr4 phosphorylation and eventually may lead to changes in transcription. Overall, we report the first case of structural and kinetic effects of phosphorylated Thr4 on CTD modifying enzymes. Our results support a model in which a combinatorial cascade of CTD modification can modulate transcription.
Organizational Affiliation:
Department of Chemistry and Biochemistry and ‡Institute for Cellular and Molecular Biology, University of Texas at Austin , Austin, Texas 78712, United States.