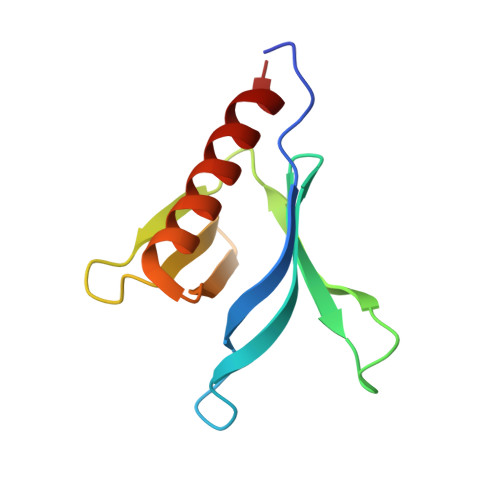
Crystal Structure of the Pleckstrin Homology Domain from the Ceramide Transfer Protein: Implications for Conformational Change upon Ligand Binding.
Prashek, J., Truong, T., Yao, X.(2013) PLoS One 8: e79590-e79590
- PubMed: 24260258
- DOI: https://doi.org/10.1371/journal.pone.0079590
- Primary Citation of Related Structures:
4HHV - PubMed Abstract:
Ceramide transfer protein (CERT) is responsible for the nonvesicular trafficking of ceramide from the endoplasmic reticulum (ER) to the trans Golgi network where it is converted to sphingomyelin (SM). The N-terminal pleckstrin homology (PH) domain is required for Golgi targeting of CERT by recognizing the phosphatidylinositol 4-phosphate (PtdIns(4)P) enriched in the Golgi membrane. We report a crystal structure of the CERT PH domain. This structure contains a sulfate that is hydrogen bonded with residues in the canonical ligand-binding pocket of PH domains. Our nuclear magnetic resonance (NMR) chemical shift perturbation (CSP) analyses show sulfate association with CERT PH protein resembles that of PtdIns(4)P, suggesting that the sulfate bound structure likely mimics the holo form of CERT PH protein. Comparison of the sulfate bound structure with the apo form solution structure shows structural rearrangements likely occur upon ligand binding, suggesting conformational flexibility in the ligand-binding pocket. This structural flexibility likely explains CERT PH domain's low affinity for PtdIns(4)P, a property that is distinct from many other PH domains that bind to their phosphoinositide ligands tightly. This unique structural feature of CERT PH domain is probably tailored towards the transfer activity of CERT protein where it needs to shuttle between ER and Golgi and therefore requires short resident time on ER and Golgi membranes.
Organizational Affiliation:
Division of Molecular Biology and Biochemistry, School of Biological Sciences, University of Missouri Kansas City, Kansas City, Missouri, United States of America.