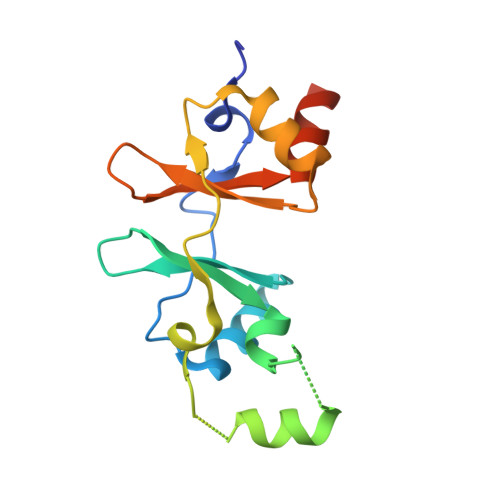
Crystal structure of CBSX1 (loop deletion)
Jeong, B.C., Park, S.H., Yoo, K.S., Shin, J.S., Song, H.K.(2012) Biochem Biophys Res Commun
- PubMed: 23159611
- DOI: https://doi.org/10.1016/j.bbrc.2012.10.139
- Primary Citation of Related Structures:
4GQV, 4GQW - PubMed Abstract:
The single cystathionine β-synthase (CBS) pair proteins from Arabidopsis thaliana have been identified as being a redox regulator of the thioredoxin (Trx) system. CBSX1 and CBSX2, which are two of the six Arabidopsis cystathione β-synthase domain-containing proteins that contain only a single CBS pair, have close sequence similarity. Recently, the crystal structure of CBSX2 was determined and a significant portion of the internal region was disordered. In this study, crystal structures of full-length CBSX1 and the internal loop deleted (Δloop) form are reported at resolutions of 2.4 and 2.2Å, respectively. The structures of CBSX1 show that they form anti-parallel dimers along their central twofold axis and have a unique ∼155° bend along the side. This is different from the angle of CBSX2, which is suggestive of the flexible nature of the relative angle between the monomers. The biochemical data that were obtained using the deletion as well as point mutants of CBSX1 confirmed the importance of AMP-ligand binding in terms of enhancing Trx activity.
Organizational Affiliation:
School of Life Sciences and Biotechnology, Korea University, Seoul 136-701, Republic of Korea.