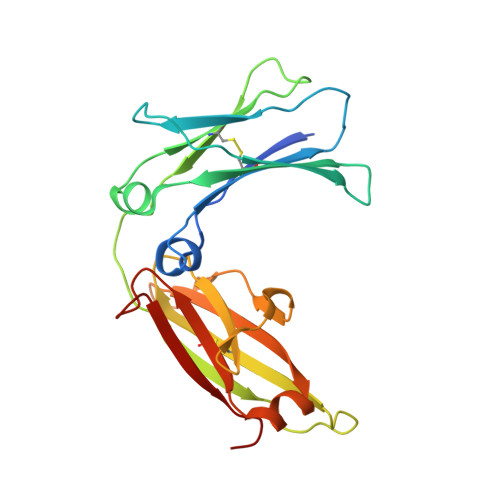
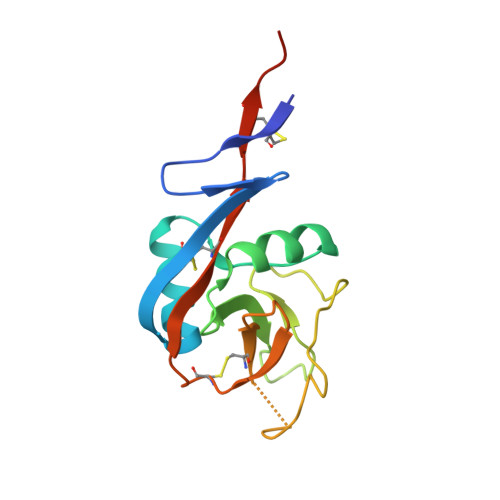
Crystal structure of IgE bound to its B-cell receptor CD23 reveals a mechanism of reciprocal allosteric inhibition with high affinity receptor Fc{varepsilon}RI.
Dhaliwal, B., Yuan, D., Pang, M.O., Henry, A.J., Cain, K., Oxbrow, A., Fabiane, S.M., Beavil, A.J., McDonnell, J.M., Gould, H.J., Sutton, B.J.(2012) Proc Natl Acad Sci U S A 109: 12686-12691
- PubMed: 22802656
- DOI: https://doi.org/10.1073/pnas.1207278109
- Primary Citation of Related Structures:
4EZM - PubMed Abstract:
The role of IgE in allergic disease mechanisms is performed principally through its interactions with two receptors, FcεRI on mast cells and basophils, and CD23 (FcεRII) on B cells. The former mediates allergic hypersensitivity, the latter regulates IgE levels, and both receptors, also expressed on antigen-presenting cells, contribute to allergen uptake and presentation to the immune system. We have solved the crystal structure of the soluble lectin-like "head" domain of CD23 (derCD23) bound to a subfragment of IgE-Fc consisting of the dimer of Cε3 and Cε4 domains (Fcε3-4). One CD23 head binds to each heavy chain at the interface between the two domains, explaining the known 2:1 stoichiometry and suggesting mechanisms for cross-linking membrane-bound trimeric CD23 by IgE, or membrane IgE by soluble trimeric forms of CD23, both of which may contribute to the regulation of IgE synthesis by B cells. The two symmetrically located binding sites are distant from the single FcεRI binding site, which lies at the opposite ends of the Cε3 domains. Structural comparisons with both free IgE-Fc and its FcεRI complex reveal not only that the conformational changes in IgE-Fc required for CD23 binding are incompatible with FcεRI binding, but also that the converse is true. The two binding sites are allosterically linked. We demonstrate experimentally the reciprocal inhibition of CD23 and FcεRI binding in solution and suggest that the mutual exclusion of receptor binding allows IgE to function independently through its two receptors.
Organizational Affiliation:
Randall Division of Cell and Molecular Biophysics, King's College London, London SE1 1UL, United Kingdom.