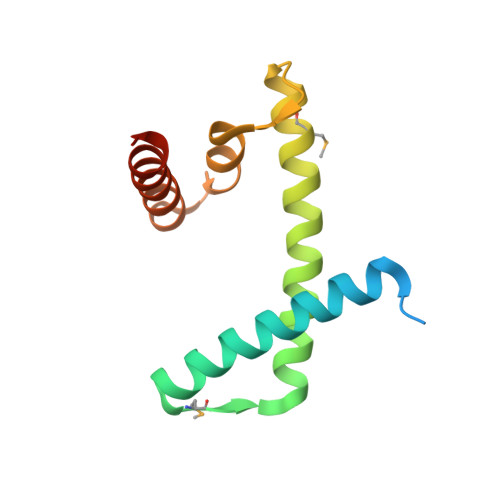
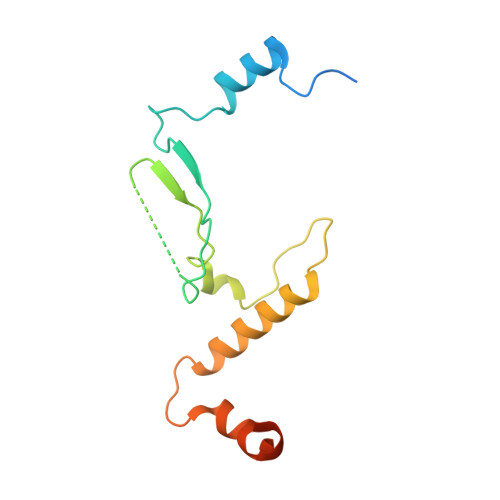
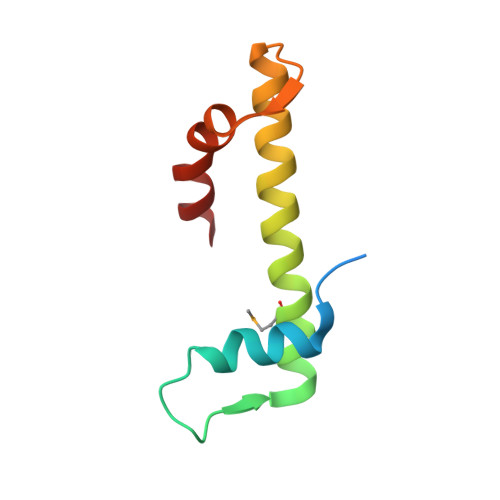
The structure of the FANCM-MHF complex reveals physical features for functional assembly
Tao, Y., Jin, C., Li, X., Qi, S., Chu, L., Niu, L., Yao, X., Teng, M.(2012) Nat Commun 3: 782-782
- PubMed: 22510687
- DOI: https://doi.org/10.1038/ncomms1779
- Primary Citation of Related Structures:
4DRA, 4DRB - PubMed Abstract:
Fanconi anaemia is a rare genetic disease characterized by chromosomal instability and cancer susceptibility. The Fanconi anaemia complementation group protein M (FANCM) forms an evolutionarily conserved DNA-processing complex with MHF1/MHF2 (histone-fold-containing proteins), which is essential for DNA repair in response to genotoxic stress. Here we present the crystal structures of the MHF1-MHF2 complex alone and bound to a fragment of FANCM (FANCM(661-800), designated FANCM-F). The structures show that MHF1 and MHF2 form a compact tetramer to which FANCM-F binds through a 'dual-V' shaped structure. FANCM-F and (MHF1-MHF2)(2) cooperate to constitute a new DNA-binding site that is coupled to the canonical L1L2 region. Perturbation of the MHF-FANCM-F structural plasticity changes the localization of FANCM in vivo. The MHF-FANCM interaction and its subcellular localization are altered by a disease-associated mutant of FANCM. These findings reveal the molecular basis of MHF-FANCM recognition and provide mechanistic insights into the pathway leading to Fanconi anaemia.
Organizational Affiliation:
Key Laboratory of Structural Biology, Chinese Academy of Sciences, Hefei, Anhui 230026, China.