Cc2D1A is a Regulator of Escrt-III Chmp4B.
Martinelli, N., Hartlieb, B., Usami, Y., Sabin, C., Dordor, A., Miguet, N., Avilov, S.V., Ribeiro, E.A., Gottlinger, H., Weissenhorn, W.(2012) J Mol Biol 419: 75
- PubMed: 22406677
- DOI: https://doi.org/10.1016/j.jmb.2012.02.044
- Primary Citation of Related Structures:
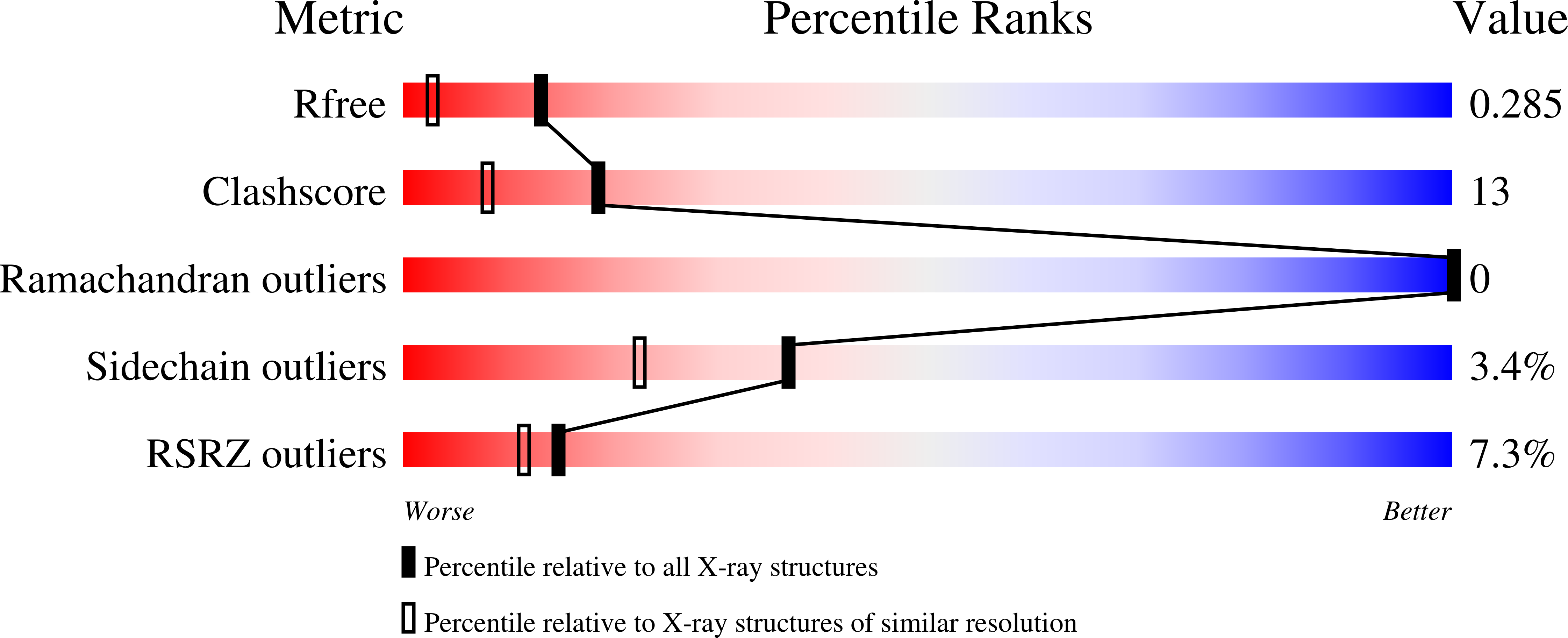
4ABM - PubMed Abstract:
Endosomal sorting complexes required for transport (ESCRTs) regulate diverse processes ranging from receptor sorting at endosomes to distinct steps in cell division and budding of some enveloped viruses. Common to all processes is the membrane recruitment of ESCRT-III that leads to membrane fission. Here, we show that CC2D1A is a novel regulator of ESCRT-III CHMP4B function. We demonstrate that CHMP4B interacts directly with CC2D1A and CC2D1B with nanomolar affinity by forming a 1:1 complex. Deletion mapping revealed a minimal CC2D1A-CHMP4B binding construct, which includes a short linear sequence within the third DM14 domain of CC2D1A. The CC2D1A binding site on CHMP4B was mapped to the N-terminal helical hairpin. Based on a crystal structure of the CHMP4B helical hairpin, two surface patches were identified that interfere with CC2D1A interaction as determined by surface plasmon resonance. Introducing these mutations into a C-terminal truncation of CHMP4B that exerts a potent dominant negative effect on human immunodeficiency virus type 1 budding revealed that one of the mutants lost this effect completely. This suggests that the identified CC2D1A binding surface might be required for CHMP4B polymerization, which is consistent with the finding that CC2D1A binding to CHMP4B prevents CHMP4B polymerization in vitro. Thus, CC2D1A might act as a negative regulator of CHMP4B function.
Organizational Affiliation:
Unit of Virus Host Cell Interactions UMI 3265, Université Joseph Fourier-EMBL-CNRS, 6 rue Jules Horowitz, 38042 Grenoble Cedex 9, France.