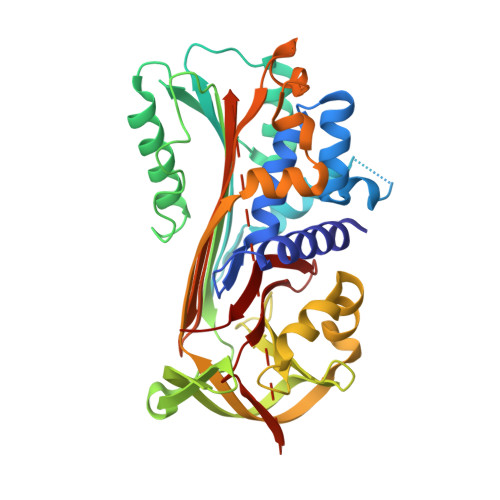
Psoriasis pathogenesis - Pso p27 constitutes a compact structure forming large aggregates.
Lysvand, H., Helland, R., Hagen, L., Slupphaug, G., Iversen, O.J.(2015) Biochem Biophys Rep 2: 132-136
- PubMed: 29124154
- DOI: https://doi.org/10.1016/j.bbrep.2015.06.001
- Primary Citation of Related Structures:
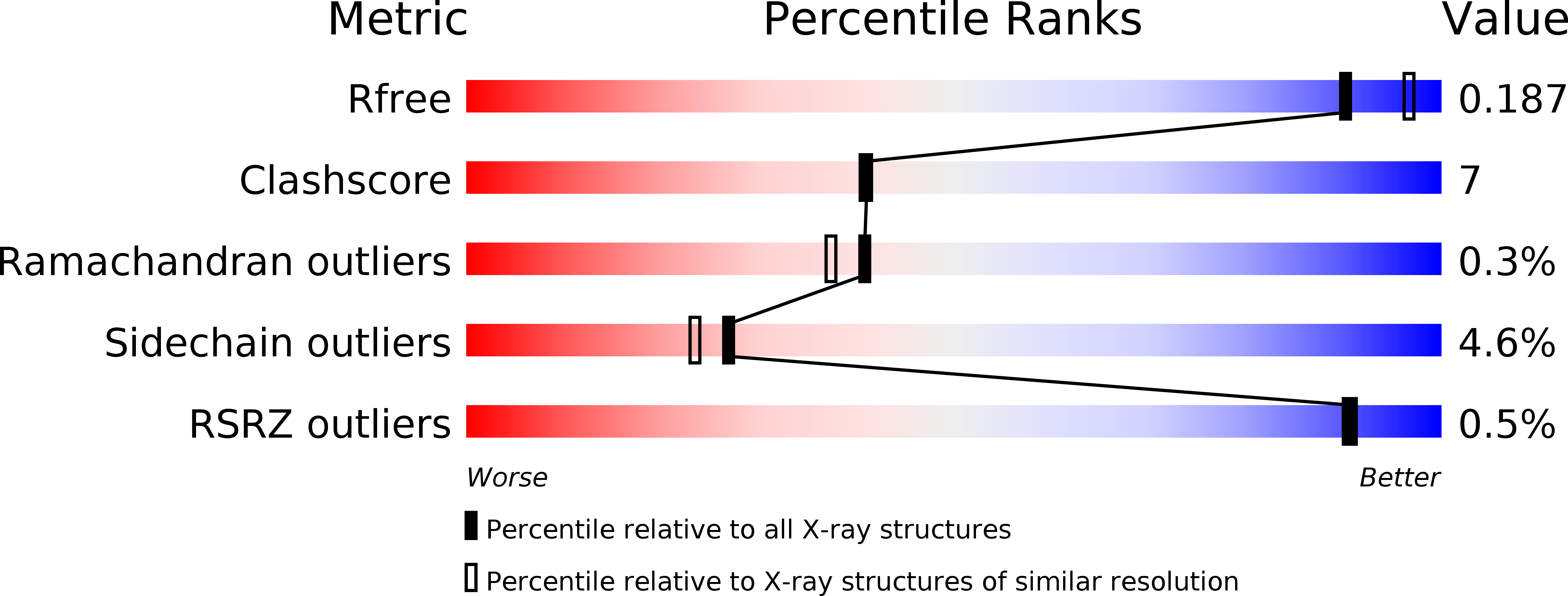
4ZK0, 4ZK3 - PubMed Abstract:
Psoriasis is a chronic inflammatory skin disease. The absence of microbial organisms as potential causal agents has given rise to the hypothesis that the inflammation is due to an autoimmune reaction. The defined inflamed areas of the skin lesions argue for an immunological disease with a local production of a causal antigen. Pso p27 is a protein generated in mast cells in psoriatic plaques, but not in uninvolved skin. We recently demonstrated that the Pso p27 is generated by cleavage of SerpinB3 (SCCA1) in the presence of mast cell associated chymase. In this communication we demonstrate by X-ray crystallographic analysis that the cleavage products associate into a complex similar to SCCA1, but with the reactive centre loop inserted into a 5-stranded central β-sheet. Native gel electrophoresis show that these Pso p27 complexes form large aggregates which may be of significance with respect to an immunogenic role of Pso p27.
Organizational Affiliation:
Department of Laboratory Medicine, Children's and Women's Health, Faculty of Medicine, Norwegian University of Science and Technology, NTNU, Postbox 8905, N-7491 Trondheim, Norway.