An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.
Leung, D.W., Borek, D., Luthra, P., Binning, J.M., Anantpadma, M., Liu, G., Harvey, I.B., Su, Z., Endlich-Frazier, A., Pan, J., Shabman, R.S., Chiu, W., Davey, R.A., Otwinowski, Z., Basler, C.F., Amarasinghe, G.K.(2015) Cell Rep 11: 376-389
- PubMed: 25865894
- DOI: https://doi.org/10.1016/j.celrep.2015.03.034
- Primary Citation of Related Structures:
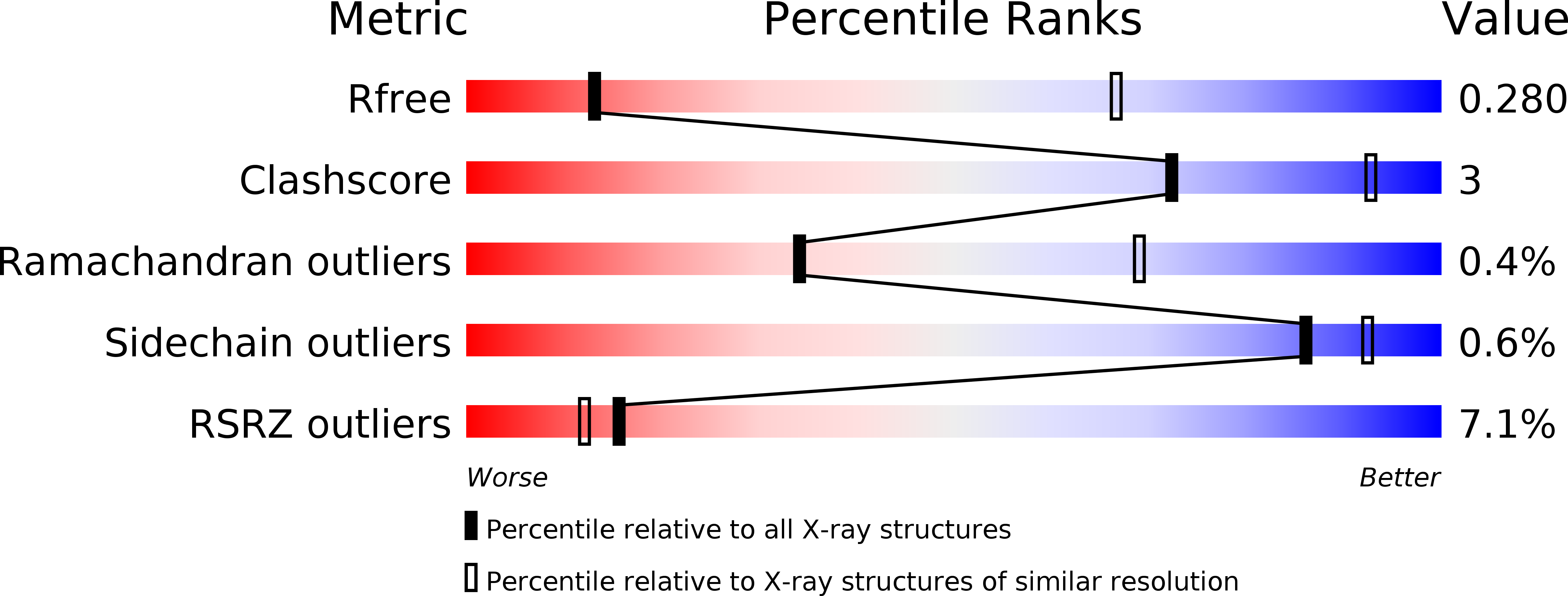
4YPI - PubMed Abstract:
During viral RNA synthesis, Ebola virus (EBOV) nucleoprotein (NP) alternates between an RNA-template-bound form and a template-free form to provide the viral polymerase access to the RNA template. In addition, newly synthesized NP must be prevented from indiscriminately binding to noncognate RNAs. Here, we investigate the molecular bases for these critical processes. We identify an intrinsically disordered peptide derived from EBOV VP35 (NPBP, residues 20-48) that binds NP with high affinity and specificity, inhibits NP oligomerization, and releases RNA from NP-RNA complexes in vitro. The structure of the NPBP/ΔNPNTD complex, solved to 3.7 Å resolution, reveals how NPBP peptide occludes a large surface area that is important for NP-NP and NP-RNA interactions and for viral RNA synthesis. Together, our results identify a highly conserved viral interface that is important for EBOV replication and can be targeted for therapeutic development.
Organizational Affiliation:
Department of Pathology and Immunology, Washington University School of Medicine, St Louis, MO 63110, USA.