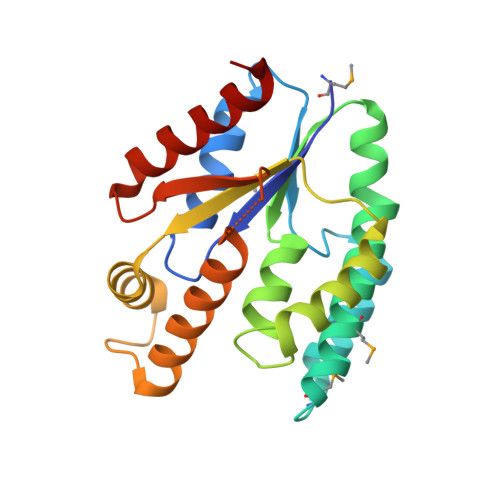
Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid
Filippova, E.V., Minasov, G., Shuvalova, L., Kiryukhina, O., Jedrzejczak, R., Babnigg, G., Rubin, E., Sacchettini, J., Joachimiak, A., Anderson, W.F.To be published.