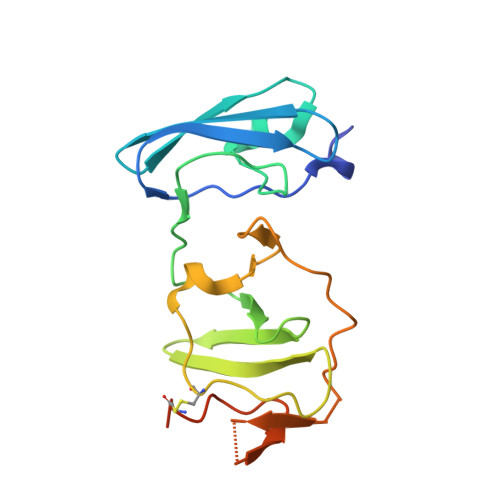
The Crystal Structure of Ns5A Domain 1 from Genotype 1A Reveals New Clues to the Mechanism of Action for Dimeric Hcv Inhibitors.
Lambert, S.M., Langley, D.R., Garnett, J.A., Angell, R., Hedgethorne, K., Meanwell, N.A., Matthews, S.J.(2014) Protein Sci 23: 723
- PubMed: 24639329
- DOI: https://doi.org/10.1002/pro.2456
- Primary Citation of Related Structures:
4CL1 - PubMed Abstract:
New direct acting antivirals (DAAs) such as daclatasvir (DCV; BMS-790052), which target NS5A function with picomolar potency, are showing promise in clinical trials. The exact nature of how these compounds have an inhibitory effect on HCV is unknown; however, major resistance mutations appear in the N-terminal region of NS5A that include the amphipathic helix and domain 1. The dimeric symmetry of these compounds suggests that they act on a dimer of NS5A, which is also consistent with the presence of dimers in crystals of NS5A domain 1 from genotype 1b. Genotype 1a HCV is less potently affected by these compounds and resistance mutations have a greater effect than in the 1b genotypes. We have obtained crystals of domain 1 of the important 1a NS5A homologue and intriguingly, our X-ray crystal structure reveals two new dimeric forms of this domain. Furthermore, the high solvent content (75%) makes it ideal for ligand-soaking. Daclatasvir (DCV) shows twofold symmetry suggesting NS5A dimers may be of physiological importance and serve as potential binding sites for DCV. These dimers also allow for new conformations of a NS5A expansive network which could explain its operation on the membranous web. Additionally, sulfates bound in the crystal structure may provide evidence for the previously proposed RNA binding groove, or explain regulation of NS5A domain 2 and 3 function and phosphorylation, by domain 1.
Organizational Affiliation:
Department of Biological Sciences, Centre for Structural Biology, Imperial College London, South Kensington, London, SW7 2AZ, United Kingdom.