Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin.
Amrich, C.G., Davis, C.P., Rogal, W.P., Shirra, M.K., Heroux, A., Gardner, R.G., Arndt, K.M., VanDemark, A.P.(2012) J Biol Chem 287: 10863-10875
- PubMed: 22318720
- DOI: https://doi.org/10.1074/jbc.M111.325647
- Primary Citation of Related Structures:
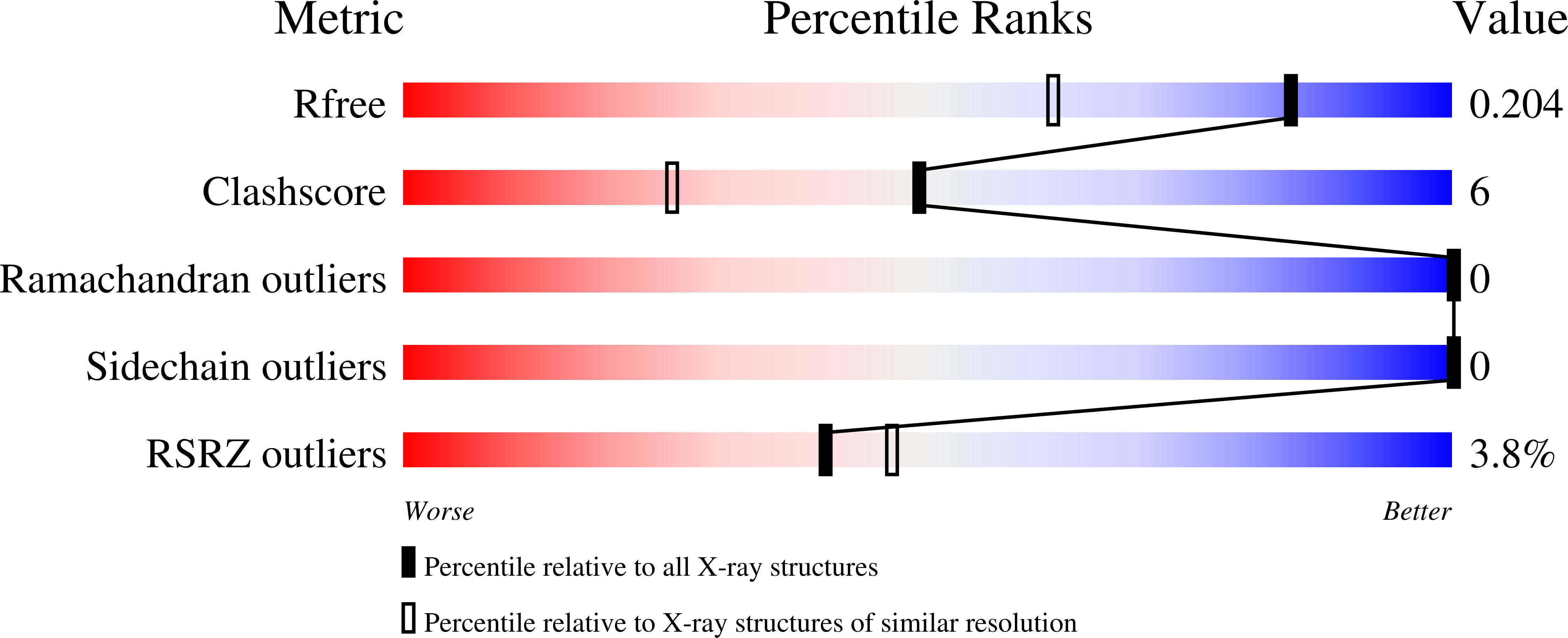
3V46 - PubMed Abstract:
The conserved Paf1 complex localizes to the coding regions of genes and facilitates multiple processes during transcription elongation, including the regulation of histone modifications. However, the mechanisms that govern Paf1 complex recruitment to active genes are undefined. Here we describe a previously unrecognized domain within the Cdc73 subunit of the Paf1 complex, the Cdc73 C-domain, and demonstrate its importance for Paf1 complex occupancy on transcribed chromatin. Deletion of the C-domain causes phenotypes associated with elongation defects without an apparent loss of complex integrity. Simultaneous mutation of the C-domain and another subunit of the Paf1 complex, Rtf1, causes enhanced mutant phenotypes and loss of histone H3 lysine 36 trimethylation. The crystal structure of the C-domain reveals unexpected similarity to the Ras family of small GTPases. Instead of a deep nucleotide-binding pocket, the C-domain contains a large but comparatively flat surface of highly conserved residues, devoid of ligand. Deletion of the C-domain results in reduced chromatin association for multiple Paf1 complex subunits. We conclude that the Cdc73 C-domain probably constitutes a protein interaction surface that functions with Rtf1 in coupling the Paf1 complex to the RNA polymerase II elongation machinery.
Organizational Affiliation:
Department of Biological Sciences, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, USA.