Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA.
Maiti, A., Noon, M.S., Mackerell, A.D., Pozharski, E., Drohat, A.C.(2012) Proc Natl Acad Sci U S A 109: 8091-8096
- PubMed: 22573813
- DOI: https://doi.org/10.1073/pnas.1201010109
- Primary Citation of Related Structures:
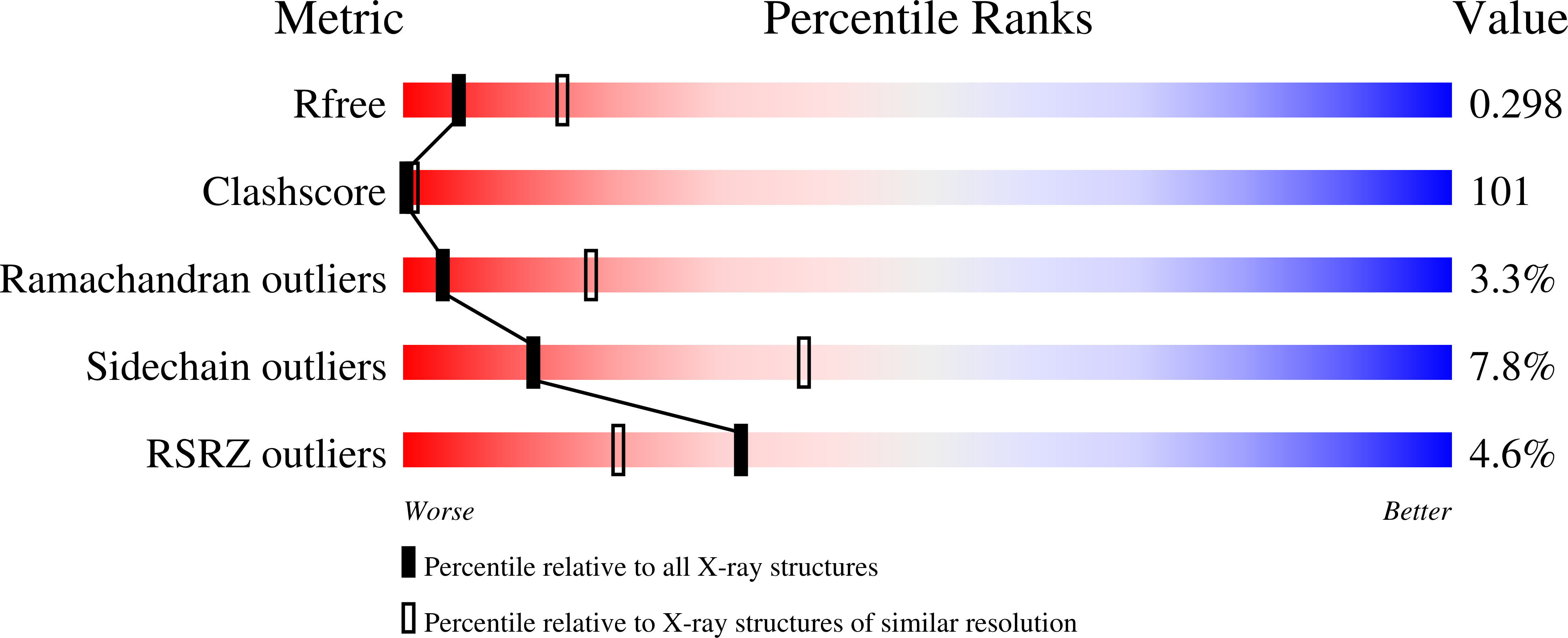
3UFJ - PubMed Abstract:
DNA base excision repair is essential for maintaining genomic integrity and for active DNA demethylation, a central element of epigenetic regulation. A key player is thymine DNA glycosylase (TDG), which excises thymine from mutagenic G·T mispairs that arise by deamination of 5-methylcytosine (mC). TDG also removes 5-formylcytosine and 5-carboxylcytosine, oxidized forms of mC produced by Tet enzymes. Recent studies show that the glycosylase activity of TDG is essential for active DNA demethylation and for embryonic development. Our understanding of how repair enzymes excise modified bases without acting on undamaged DNA remains incomplete, particularly for mismatch glycosylases such as TDG. We solved a crystal structure of TDG (catalytic domain) bound to a substrate analog and characterized active-site residues by mutagenesis, kinetics, and molecular dynamics simulations. The studies reveal how TDG binds and positions the nucleophile (water) and uncover a previously unrecognized catalytic residue (Thr197). Remarkably, mutation of two active-site residues (Ala145 and His151) causes a dramatic enhancement in G·T glycosylase activity but confers even greater increases in the aberrant removal of thymine from normal A·T base pairs. The strict conservation of these residues may reflect a mechanism used to strike a tolerable balance between the requirement for efficient repair of G·T lesions and the need to minimize aberrant action on undamaged DNA, which can be mutagenic and cytotoxic. Such a compromise in G·T activity can account in part for the relatively weak G·T activity of TDG, a trait that could potentially contribute to the hypermutability of CpG sites in cancer and genetic disease.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, School of Medicine, University of Maryland, Baltimore, MD 21201, USA.