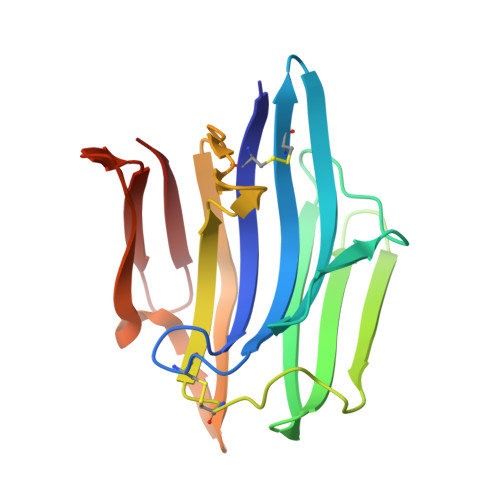
The crystal structure of an intermediate dimer of aspergilloglutamic peptidase that mimics the enzyme-activation product complex produced upon autoproteolysis.
Sasaki, H., Kubota, K., Lee, W.C., Ohtsuka, J., Kojima, M., Iwata, S., Nakagawa, A., Takahashi, K., Tanokura, M.(2012) J Biochem 152: 45-52
- PubMed: 22569035
- DOI: https://doi.org/10.1093/jb/mvs050
- Primary Citation of Related Structures:
3TRS - PubMed Abstract:
Aspergilloglutamic peptidase from Aspergillus niger var. macrosporus (AGP) is one of the so-called pepstatin-insensitive acid endopeptidases, which are distinct from the well-studied aspartic peptidases. Among the known homologues of the glutamic peptidases, AGP is a unique two-chain enzyme with a light chain and a heavy chain bound non-covalently with each other, and thus is an interesting target for protein structure-function relationship studies. In this article, we report the crystal structure of a dimeric form of the enzyme at a resolution of 1.6 Å. This form has a unique structure in which the C-terminal region of the light chain of one of the molecules binds to the active site cleft of the other molecule like a part of a substrate. This form mimics the enzyme-activation product complex produced upon autoproteolysis, and provides a structural clue that could help to clarify the activation mechanism. This type of dimeric structure of a peptidase is here reported for the first time.
Organizational Affiliation:
Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Tokyo 113-0033, Japan.