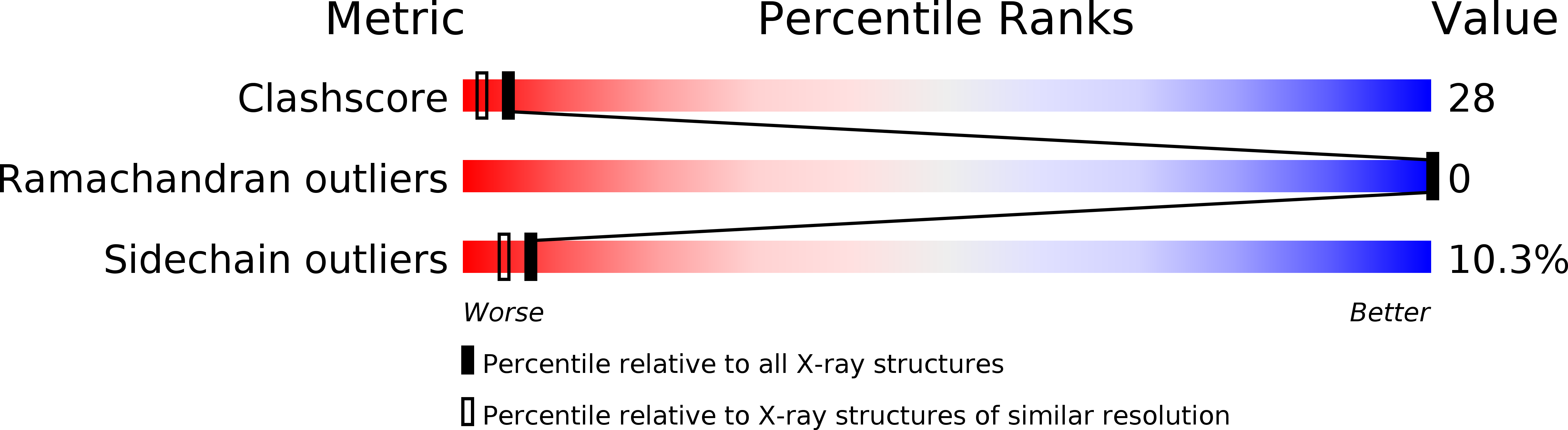Changes in crystallographic structure and thermostability of a Cu,Zn superoxide dismutase mutant resulting from the removal of a buried cysteine.
McRee, D.E., Redford, S.M., Getzoff, E.D., Lepock, J.R., Hallewell, R.A., Tainer, J.A.(1990) J Biol Chem 265: 14234-14241
- PubMed: 2387847
- DOI: https://doi.org/10.2210/pdb3sod/pdb
- Primary Citation of Related Structures:
3SOD - PubMed Abstract:
In principle, protein thermostability depends on efficient interior packing of apolar residues and on avoidance of irreversible denaturation in the unfolded state. To study these effects, the single free cysteine in the highly stable enzyme bovine Cu,Zn superoxide dismutase was mutated to alanine (Cys6----Ala), and the recombinant protein was expressed in yeast, purified, characterized for reversible and irreversible denaturation, crystallized isomorphously to the wild-type enzyme, and used to determine the atomic structure. Removal of the chemically reactive thiol significantly decreased the rate of irreversible denaturation (as monitored by thermal inactivation at 70 degrees C), but the observed energetic cost (delta delta G of 0.7-1.3 kcal/mol as determined by differential scanning calorimetry) was much less than predicted from either the change in hydrophobicity or packing due to removal of the interior sulfur atom. X-ray diffraction data were collected to 2.1-A resolution using an area detector, and the atomic model for the mutant enzyme was determined by fitting to electron density difference maps, followed by reciprocal space refinement both with stereochemical restraints using PROLSQ and with molecular dynamics using X-PLOR. The refined 2.1-A resolution crystallographic structure suggests that small concerted and compensating shifts (less than 0.5 A) of the surrounding side chains and of the adjacent N- and C-terminal beta-strands significantly reduced the energetic cost of the interior mutation by improving packing and stereochemistry in the mutant enzyme. Taken together, these results differentiate between the effects of reversible and irreversible processes as they impact the design of thermostable proteins and suggest that relatively subtle concerted shifts can significantly reduce the energetic cost of evolutionary variation in internal residues of proteins with Greek key beta-barrel folds.
Organizational Affiliation:
Department of Molecular Biology, Research Institute of Scripps Clinic, La Jolla, California 92037.