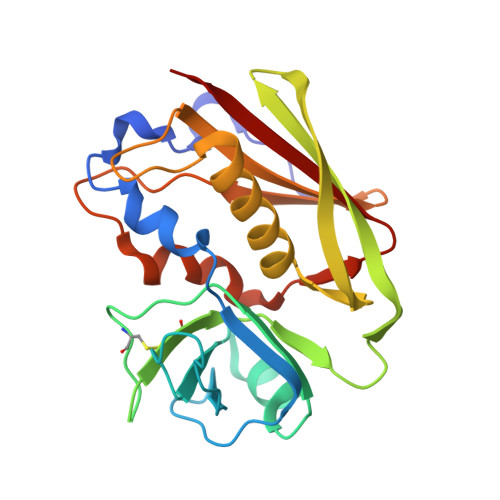
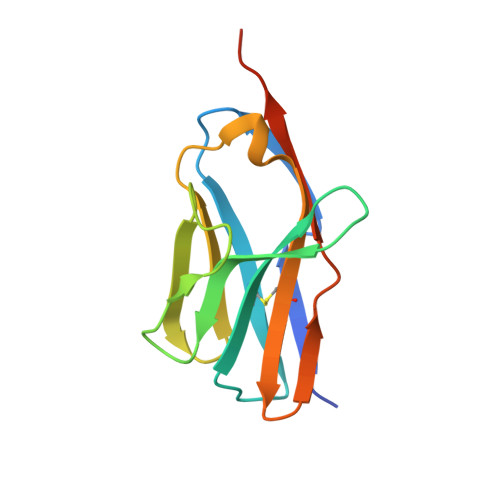
Molecular basis of a million-fold affinity maturation process in a protein-protein interaction.
Bonsor, D.A., Postel, S., Pierce, B.G., Wang, N., Zhu, P., Buonpane, R.A., Weng, Z., Kranz, D.M., Sundberg, E.J.(2011) J Mol Biol 411: 321-328
- PubMed: 21689661
- DOI: https://doi.org/10.1016/j.jmb.2011.06.009
- Primary Citation of Related Structures:
3R8B - PubMed Abstract:
Protein engineering is becoming increasingly important for pharmaceutical applications where controlling the specificity and affinity of engineered proteins is required to create targeted protein therapeutics. Affinity increases of several thousand-fold are now routine for a variety of protein engineering approaches, and the structural and energetic bases of affinity maturation have been investigated in a number of such cases. Previously, a 3-million-fold affinity maturation process was achieved in a protein-protein interaction composed of a variant T-cell receptor fragment and a bacterial superantigen. Here, we present the molecular basis of this affinity increase. Using X-ray crystallography, shotgun reversion/replacement scanning mutagenesis, and computational analysis, we describe, in molecular detail, a process by which extrainterfacial regions of a protein complex can be rationally manipulated to significantly improve protein engineering outcomes.
Organizational Affiliation:
Boston Biomedical Research Institute, 64 Grove Street, Watertown, MA 02472, USA.