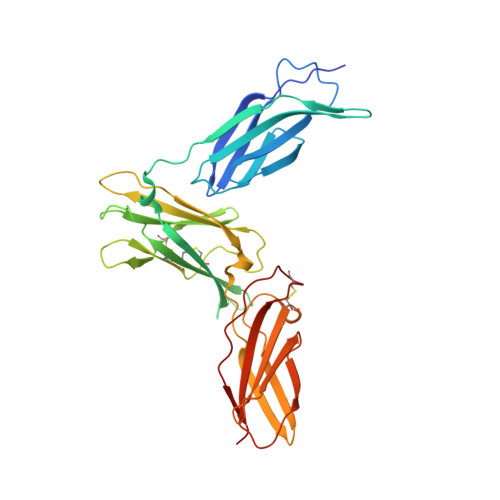
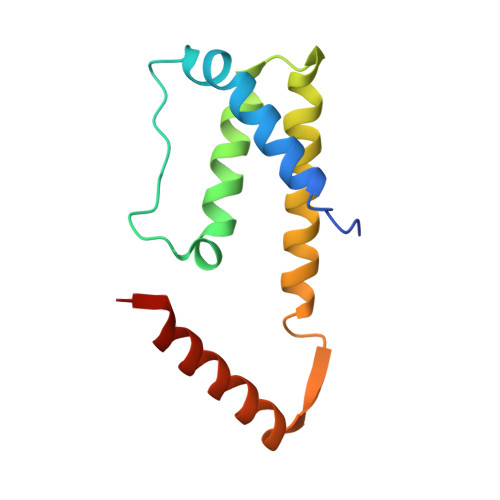
Structure Analysis of the IL-5 Ligand-Receptor Complex Reveals a Wrench-like Architecture for IL-5Ralpha.
Patino, E., Kotzsch, A., Saremba, S., Nickel, J., Schmitz, W., Sebald, W., Mueller, T.D.(2011) Structure 19: 1864-1875
- PubMed: 22153509
- DOI: https://doi.org/10.1016/j.str.2011.08.015
- Primary Citation of Related Structures:
3QT2 - PubMed Abstract:
Interleukin-5 (IL-5) is the key mediator for the function of eosinophil granulocytes, whose deregulation is characteristic of hypereosinophilic diseases and presumably contributes to allergic asthma. IL-5 signaling involves two transmembrane receptors, IL-5Rα and the common β chain, which upon formation of the ternary complex activate the JAK/STAT signaling cascade. To investigate the mechanism underlying ligand-receptor recognition, we determined the structure of IL-5 bound to the extracellular domain of IL-5Rα. IL-5 makes contact with all three fibronectin III-like domains of IL-5Rα, with the receptor architecture resembling a wrench. Mutagenesis data provide evidence that this wrench-like architecture is likely preformed. The structure demonstrates that for steric reasons, homodimeric IL-5 can bind only one receptor molecule, even though two equivalent receptor-binding sites exist. In regard to strong efforts being made to develop IL-5 antagonists for treating asthma and hypereosinophilic diseases, the advances in molecular understanding provided by this structure are of greatest value.
Organizational Affiliation:
Lehrstuhl für Molekulare Pflanzenphysiologie und Biophysik, Julius-von-Sachs Institut der Universität Würzburg, Julius-von-Sachs Platz 2, D-97082 Würzburg, Germany.