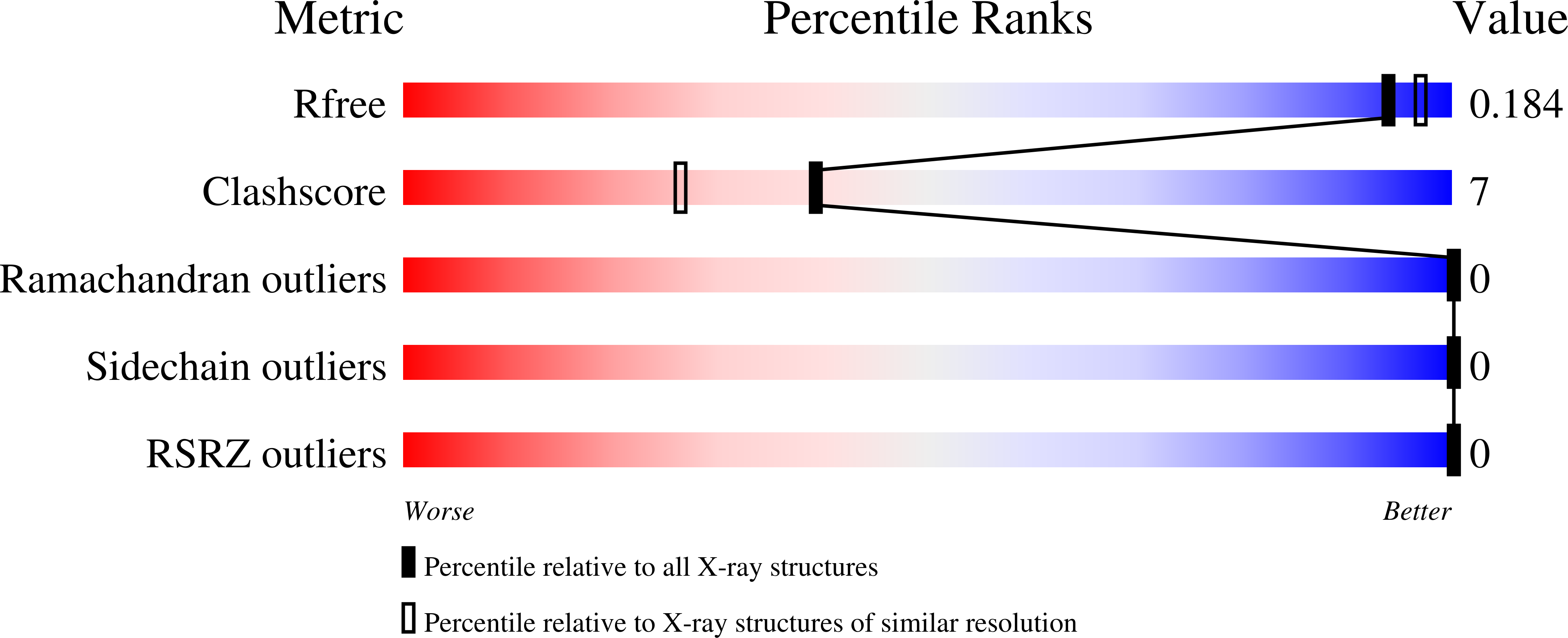
An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase
Lai, K.K., Stogios, P.J., Vu, C., Xu, X., Cui, H., Molloy, S., Savchenko, A., Yakunin, A., Gonzalez, C.F.(2011) PLoS One 6: e23269-e23269
- PubMed: 21876742
- DOI: https://doi.org/10.1371/journal.pone.0023269
- Primary Citation of Related Structures:
3PF8, 3PF9, 3PFB, 3PFC, 3QM1, 3S2Z - PubMed Abstract:
Microbial enzymes produced in the gastrointestinal tract are primarily responsible for the release and biochemical transformation of absorbable bioactive monophenols. In the present work we described the crystal structure of LJ0536, a serine cinnamoyl esterase produced by the probiotic bacterium Lactobacillus johnsonii N6.2.
Organizational Affiliation:
Department of Microbiology and Cell Science, Genetics Institute, University of Florida, Gainesville, Florida, United States of America.