Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Smith, P., Nair, P.A., Das, U., Zhu, H., Shuman, S.(2011) Nucleic Acids Res 39: 3310-3320
- PubMed: 21208981
- DOI: https://doi.org/10.1093/nar/gkq1163
- Primary Citation of Related Structures:
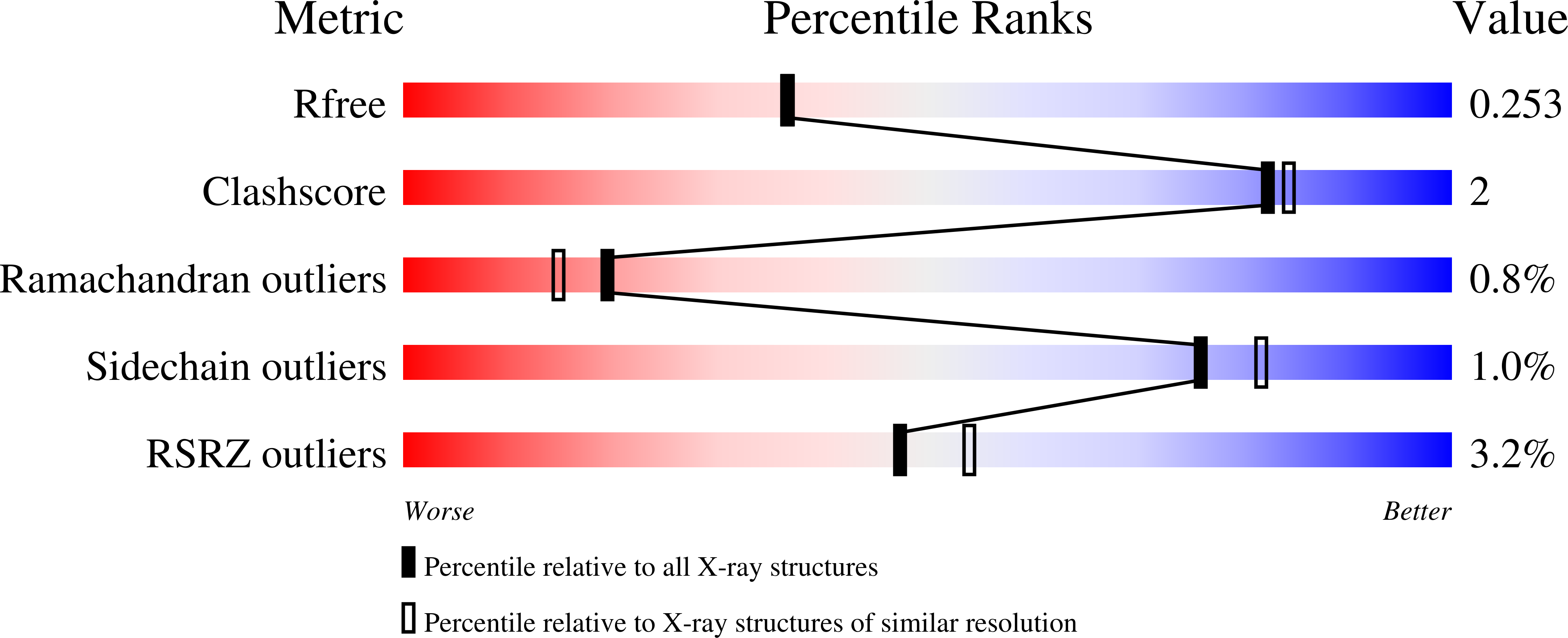
3P43, 3P4H - PubMed Abstract:
LigD 3'-phosphoesterase (PE) is a component of the bacterial NHEJ apparatus that performs 3'-end-healing reactions at DNA breaks. The tertiary structure, active site and substrate specificity of bacterial PE are unique vis-à-vis other end-healing enzymes. PE homologs are present in archaea, but their properties are uncharted. Here, we demonstrate the end-healing activities of two archaeal PEs--Candidatus Korarchaeum cryptofilum PE (CkoPE; 117 amino acids) and Methanosarcina barkeri PE (MbaPE; 151 amino acids)--and we report their atomic structures at 1.1 and 2.1 Å, respectively. Archaeal PEs are minimized versions of bacterial PE, consisting of an eight-stranded β barrel and a 3(10) helix. Their active sites are located in a crescent-shaped groove on the barrel's outer surface, wherein two histidines and an aspartate coordinate manganese in an octahedral complex that includes two waters and a phosphate anion. The phosphate is in turn coordinated by arginine and histidine side chains. The conservation of active site architecture in bacterial and archaeal PEs, and the concordant effects of active site mutations, underscore a common catalytic mechanism, entailing transition state stabilization by manganese and the phosphate-binding arginine and histidine. Our results fortify the proposal that PEs comprise a DNA repair superfamily distributed widely among taxa.
Organizational Affiliation:
Molecular Biology Program, Sloan-Kettering Institute, New York, NY 10065, USA.