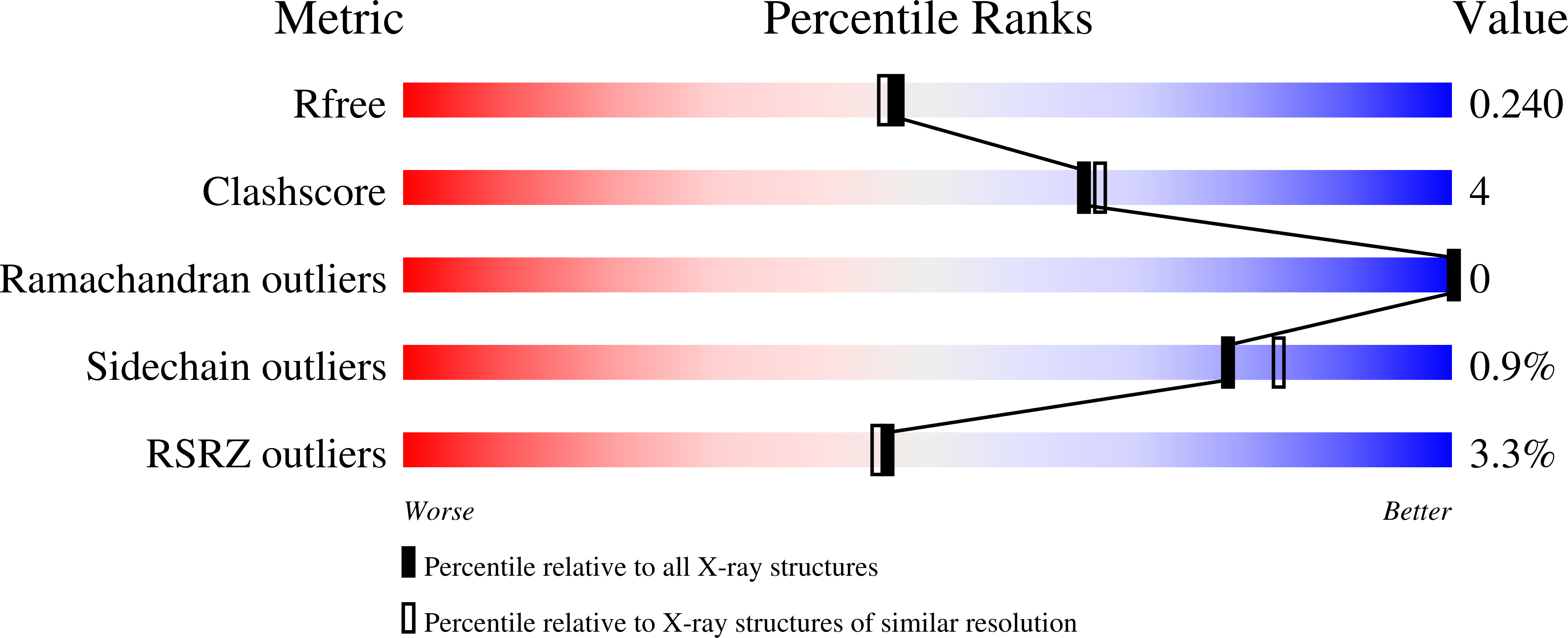
A novel structural mechanism for redox regulation of uridine phosphorylase 2 activity.
Roosild, T.P., Castronovo, S., Villoso, A., Ziemba, A., Pizzorno, G.(2011) J Struct Biol 176: 229-237
- PubMed: 21855639
- DOI: https://doi.org/10.1016/j.jsb.2011.08.002
- Primary Citation of Related Structures:
3P0E, 3P0F - PubMed Abstract:
Uridine phosphorylase (UPP) catalyzes the reversible conversion of uridine to uracil and ribose-1-phosphate and plays an important pharmacological role in activating fluoropyrimidine nucleoside chemotherapeutic agents such as 5-fluorouracil and capecitabine. Most vertebrate animals, including humans, possess two homologs of this enzyme (UPP1 & UPP2), of which UPP1 has been more thoroughly studied and is better characterized. Here, we report two crystallographic structures of human UPP2 (hUPP2) in distinctly active and inactive conformations. These structures reveal that a conditional intramolecular disulfide bridge can form within the protein that dislocates a critical phosphate-coordinating arginine residue (R100) away from the active site, disabling the enzyme. In vitro activity measurements on both recombinant hUPP2 and native mouse UPP2 confirm the redox sensitivity of this enzyme, in contrast to UPP1. Sequence analysis shows that this feature is conserved among UPP2 homologs and lacking in all UPP1 proteins due to the absence of a necessary cysteine residue. The state of the disulfide bridge has further structural consequences for one face of the enzyme that suggest UPP2 may have additional functions in sensing and initiating cellular responses to oxidative stress. The molecular details surrounding these dynamic aspects of hUPP2 structure and regulation provide new insights as to how novel inhibitors of this protein may be developed with improved specificity and affinity. As uridine is emerging as a promising protective compound in neuro-degenerative diseases, including Alzheimer's and Parkinson's, understanding the regulatory mechanisms underlying UPP control of uridine concentration is key to improving clinical outcomes in these illnesses.
Organizational Affiliation:
Department of Drug Development, Nevada Cancer Institute, One Breakthrough Way, Las Vegas, NV 89135, USA. troosild@nvcancer.org