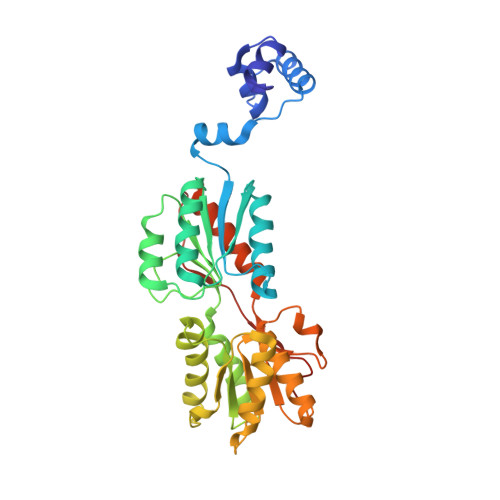
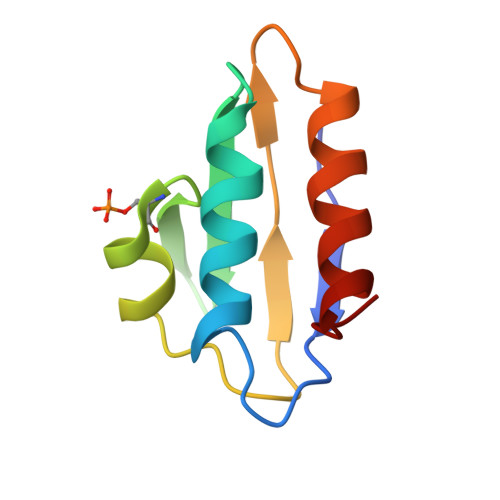
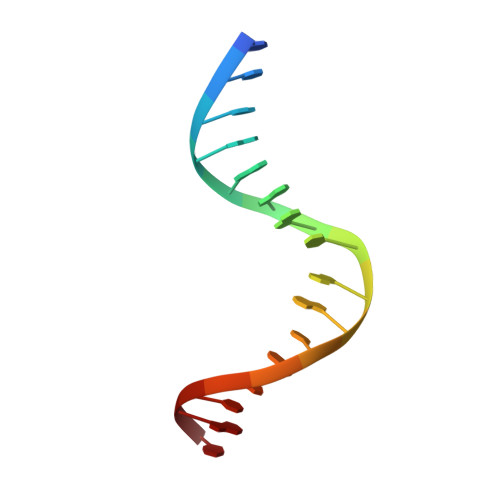
Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Schumacher, M.A., Sprehe, M., Bartholomae, M., Hillen, W., Brennan, R.G.(2011) Nucleic Acids Res 39: 2931-2942
- PubMed: 21106498
- DOI: https://doi.org/10.1093/nar/gkq1177
- Primary Citation of Related Structures:
3OQM, 3OQN, 3OQO - PubMed Abstract:
In Gram-positive bacteria, carbon catabolite protein A (CcpA) is the master regulator of carbon catabolite control, which ensures optimal energy usage under diverse conditions. Unlike other LacI-GalR proteins, CcpA is activated for DNA binding by first forming a complex with the phosphoprotein HPr-Ser46-P. Bacillus subtilis CcpA functions as both a transcription repressor and activator and binds to more than 50 operators called catabolite response elements (cres). These sites are highly degenerate with the consensus, WTGNNARCGNWWWCAW. How CcpA-(HPr-Ser46-P) binds such diverse sequences is unclear. To gain insight into this question, we solved the structures of the CcpA-(HPr-Ser46-P) complex bound to three different operators, the synthetic (syn) cre, ackA2 cre and gntR-down cre. Strikingly, the structures show that the CcpA-bound operators display different bend angles, ranging from 31° to 56°. These differences are accommodated by a flexible linkage between the CcpA helix-turn-helix-loop-helix motif and hinge helices, which allows independent docking of these DNA-binding modules. This flexibility coupled with an abundance of non-polar residues capable of non-specific nucleobase interactions permits CcpA-(HPr-Ser46-P) to bind diverse operators. Indeed, biochemical data show that CcpA-(HPr-Ser46-P) binds the three cre sites with similar affinities. Thus, the data reveal properties that license this protein to function as a global transcription regulator.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA. maschuma@mdanderson.org