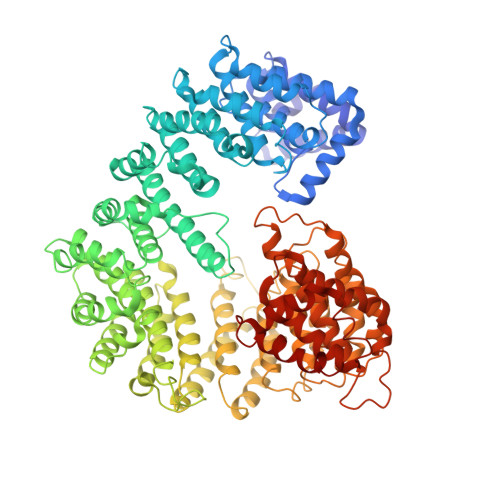
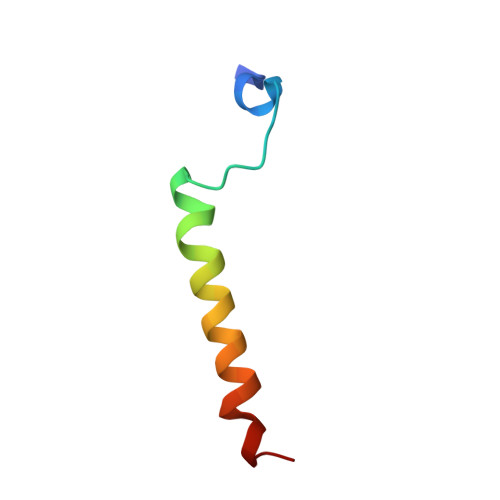
Conformational selection in the recognition of the snurportin importin beta binding domain by importin beta.
Bhardwaj, A., Cingolani, G.(2010) Biochemistry 49: 5042-5047
- PubMed: 20476751
- DOI: https://doi.org/10.1021/bi100292y
- Primary Citation of Related Structures:
3LWW - PubMed Abstract:
The structural flexibility of beta-karyopherins is critical to mediate the interaction with transport substrates, nucleoporins, and the GTPase Ran. In this paper, we provide structural evidence that the molecular recognition of the transport adaptor snurportin by importin beta follows the population selection mechanism. We have captured two drastically different conformations of importin beta bound to the snurportin importin beta binding domain trapped in the same crystallographic asymmetric unit. We propose the population selection may be a general mechanism used by beta-karyopherins to recognize transport substrates.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Thomas Jefferson University, 233 South 10th Street, Philadelphia, Pennsylvania 19107, USA.