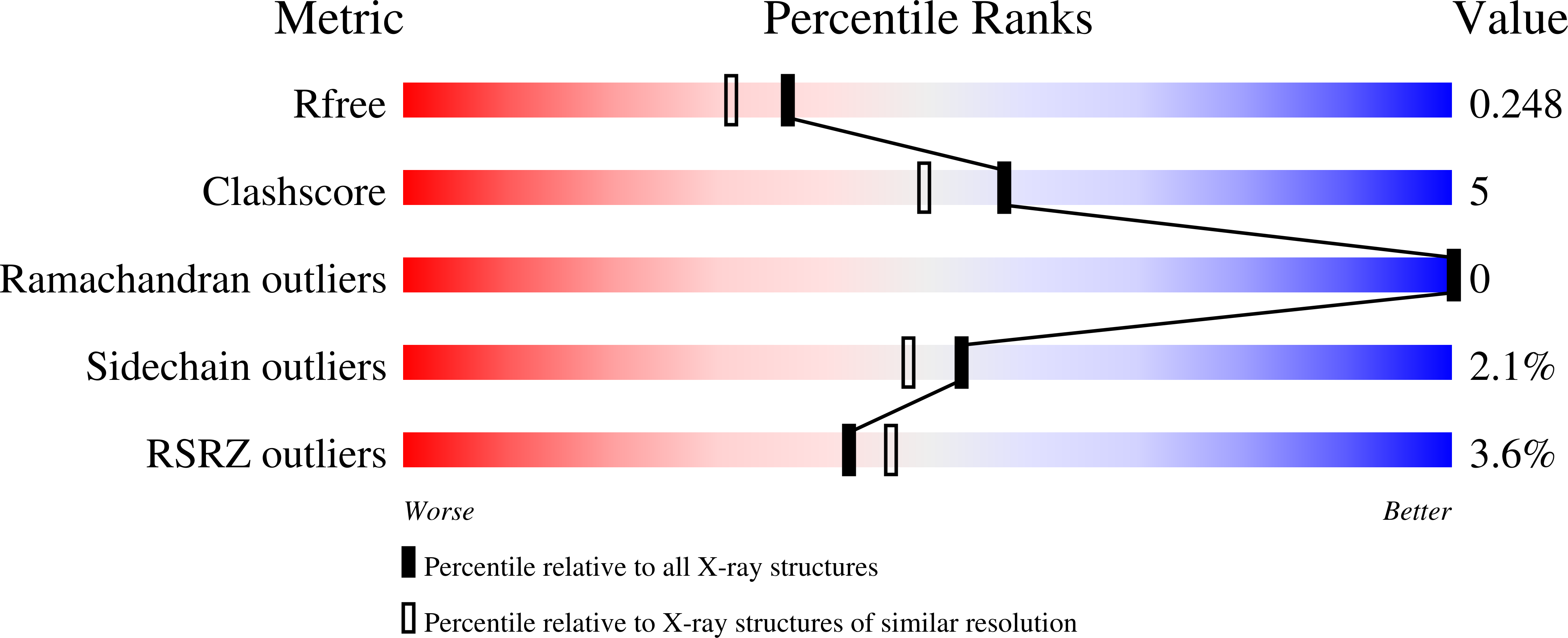
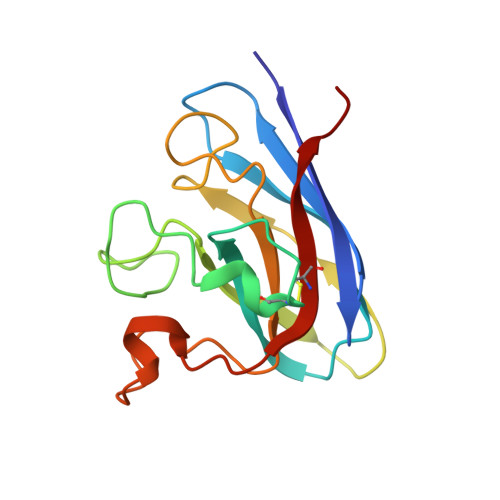
Crystal structures of holo and Cu-deficient Cu/Zn-SOD from the silkworm Bombyx mori and the implications in amyotrophic lateral sclerosis
Zhang, N.-N., He, Y.-X., Li, W.-F., Teng, Y.-B., Yu, J., Chen, Y., Zhou, C.-Z.(2010) Proteins 78: 1999-2004
- PubMed: 20310068
- DOI: https://doi.org/10.1002/prot.22709
- Primary Citation of Related Structures:
3L9E, 3L9Y
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Anhui, People's Republic of China.