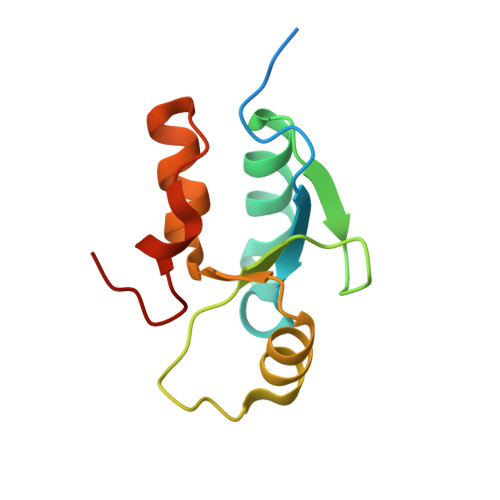
Insights from the crystal structure of the sixth BRCT domain of topoisomerase IIbeta binding protein 1.
Leung, C.C., Kellogg, E., Kuhnert, A., Hanel, F., Baker, D., Glover, J.N.(2010) Protein Sci 19: 162-167
- PubMed: 19937654
- DOI: https://doi.org/10.1002/pro.290
- Primary Citation of Related Structures:
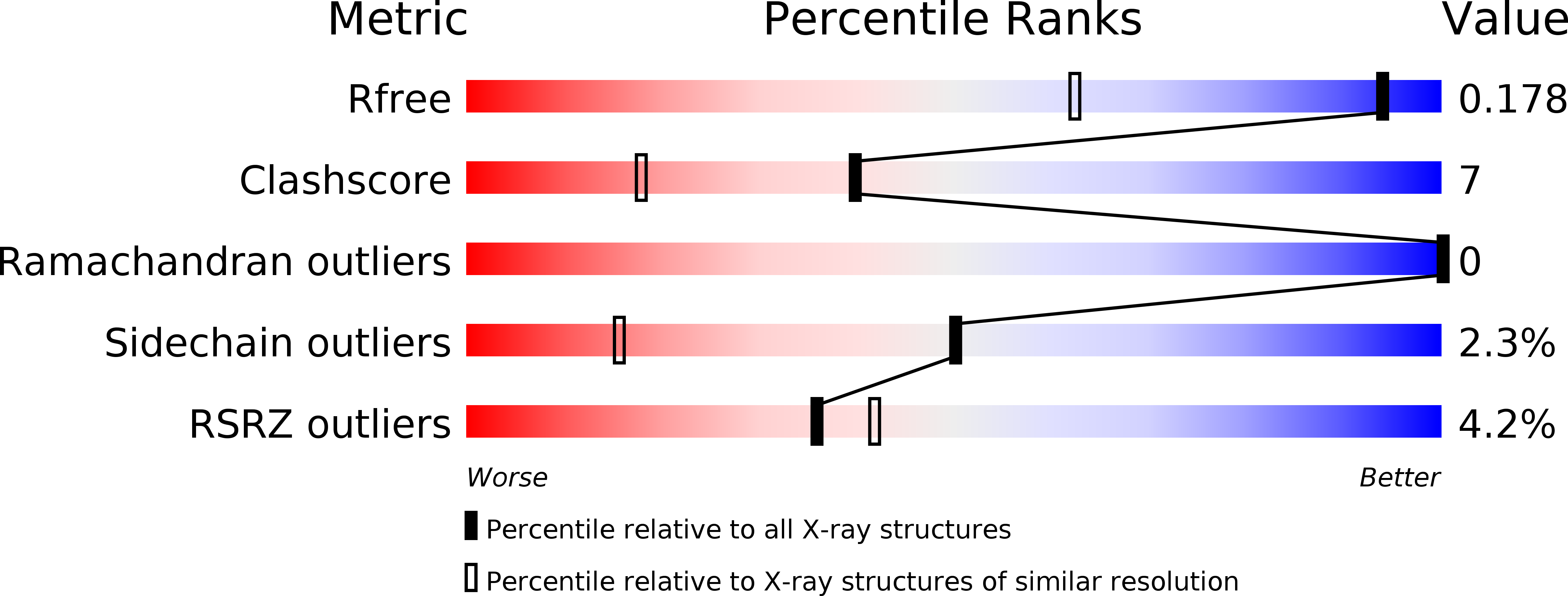
3JVE - PubMed Abstract:
Topoisomerase IIbeta binding protein 1 (TopBP1) is a major player in the DNA damage response and interacts with a number of protein partners via its eight BRCA1 carboxy-terminal (BRCT) domains. In particular, the sixth BRCT domain of TopBP1 has been implicated in binding to the phosphorylated transcription factor, E2F1, and poly(ADP-ribose) polymerase 1 (PARP-1), where the latter interaction is responsible for the poly(ADP-ribosyl)ation of TopBP1. To gain a better understanding of the nature of TopBP1 BRCT6 interactions, we solved the crystal structure of BRCT6 to 1.34 A. The crystal structure reveals a degenerate phospho-peptide binding pocket and lacks conserved hydrophobic residues involved in packing of tandem BRCT repeats, which, together with results from phospho-peptide binding studies, strongly suggest that TopBP1 BRCT6 independently does not function as a phospho-peptide binding domain. We further provide insight into poly(ADP-ribose) binding and sites of potential modification by PARP-1.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada T6G 2H7.