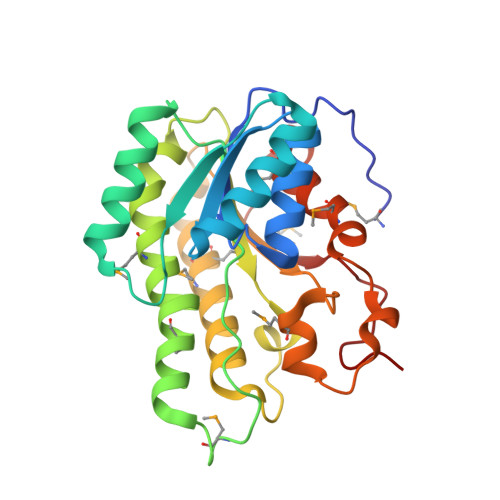
Crystal Structure of Short Chain Dehydrogenase (yciK) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 in Complex with NADP and Acetate.
Minasov, G., Halavaty, A., Skarina, T., Onopriyenko, O., Peterson, S.N., Savchenko, A., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.