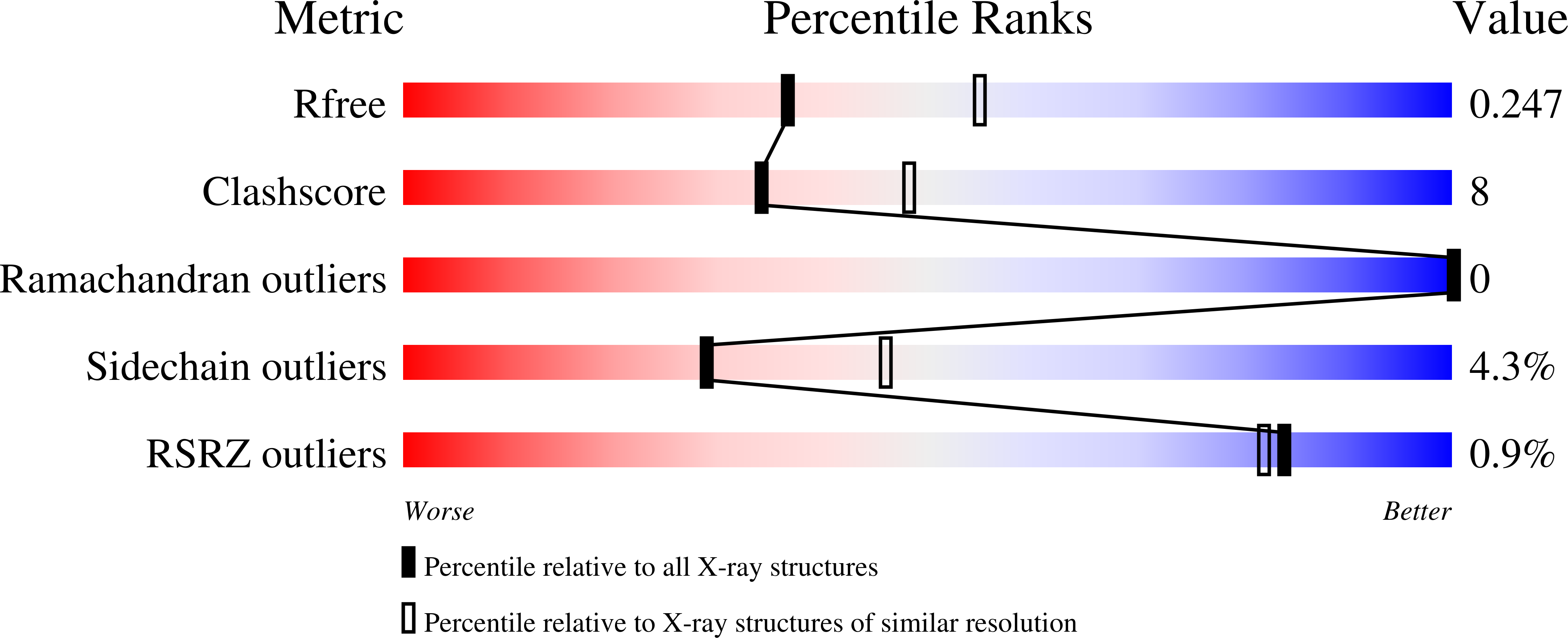
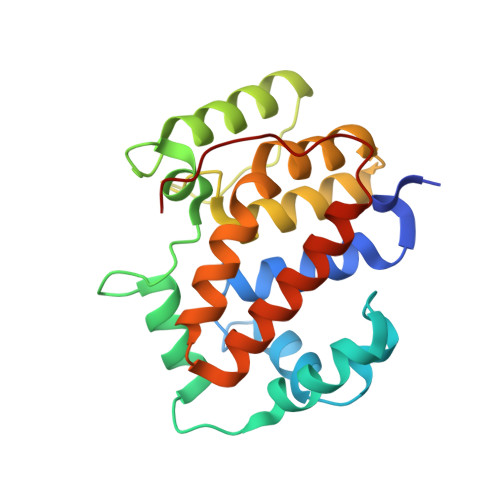
Structure of the twin-arginine signal-binding protein DmsD from Escherichia coli.
Ramasamy, S.K., Clemons, W.M.(2009) Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 746-750
- PubMed: 19652330
- DOI: https://doi.org/10.1107/S1744309109023811
- Primary Citation of Related Structures:
3CW0 - PubMed Abstract:
The translocation of folded proteins via the twin-arginine translocation (Tat) pathway is regulated to prevent the futile export of inactive substrate. DmsD is part of a class of cytoplasmic chaperones that play a role in preventing certain redox proteins from premature transport. DmsD from Escherichia coli has been crystallized in space group P4(1)2(1)2, with unit-cell parameters a = b = 97.45, c = 210.04 A, in the presence of a small peptide. The structure has been solved by molecular replacement to a resolution of 2.4 A and refined to an R factor of 19.4%. There are four molecules in the asymmetric unit that may mimic a higher order structure in vivo. There appears to be density for the peptide in a predicted binding pocket, which lends support to its role as the signal-recognition surface for this class of proteins.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125, USA.