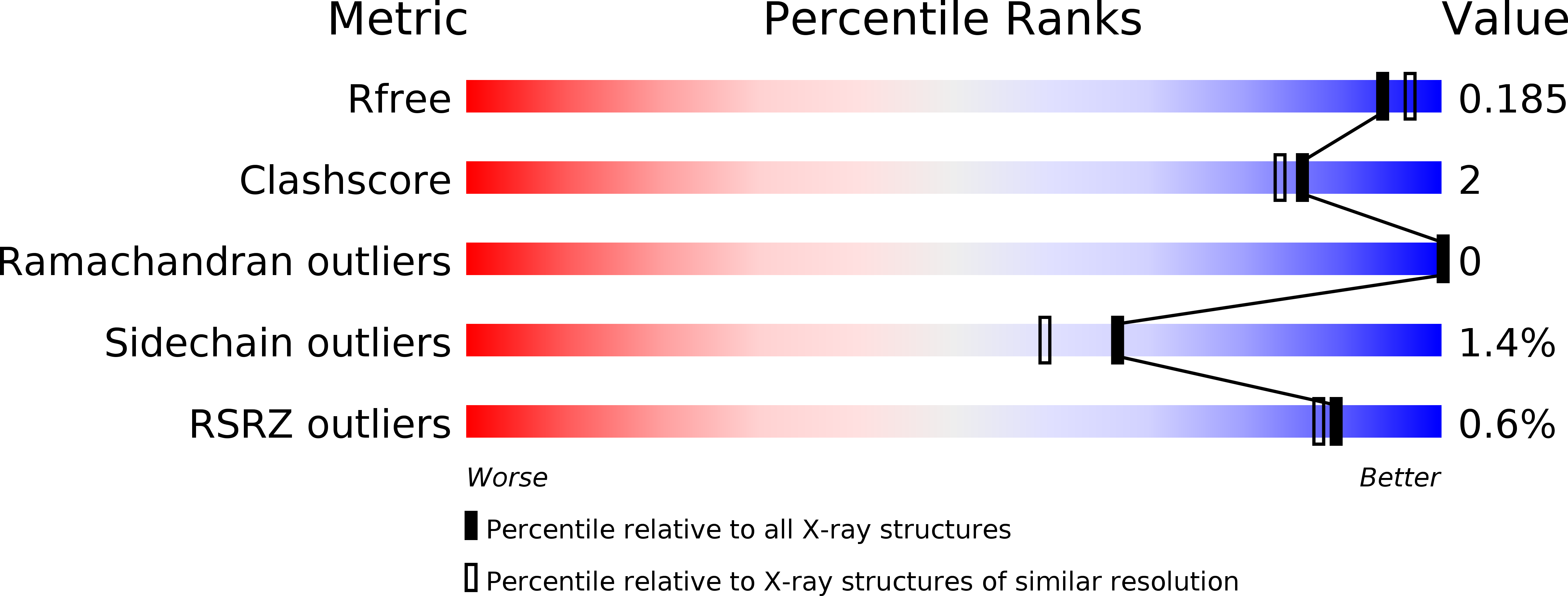
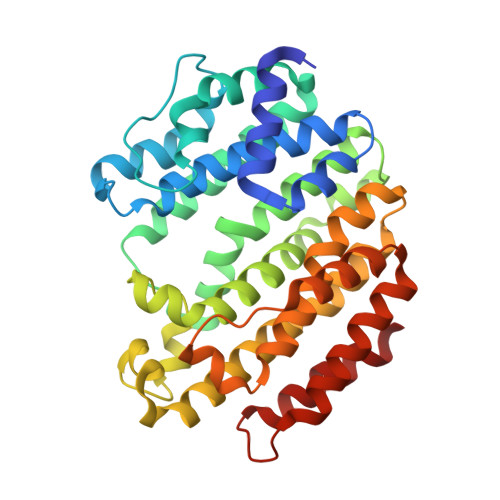
Discovery of novel tricyclic compounds as squalene synthase inhibitors
Ichikawa, M., Ohtsuka, M., Ohki, H., Haginoya, N., Itoh, M., Sugita, K., Usui, H., Suzuki, M., Terayama, K., Kanda, A.(2012) Bioorg Med Chem 20: 3072-3093
- PubMed: 22464687
- DOI: https://doi.org/10.1016/j.bmc.2012.02.054
- Primary Citation of Related Structures:
3V66 - PubMed Abstract:
In the present article, we have reported the design, synthesis, and identification of highly potent benzhydrol derivatives as squalene synthase inhibitors (compound 1). Unfortunately, the in vivo efficacies of the compounds were not enough for acquiring the clinical candidate. We continued our investigation to obtain a more in vivo efficacious template than the benzhydrol template. In our effort, we focused on a benzoxazepine ring and designed a new tricyclic scaffold by the incorporation of heterocycle into it. Prepared pyrrolobenzoxazepine derivatives showed further efficient in vitro and in vivo activities.
Organizational Affiliation:
Lead Discovery and Optimization Research Laboratories I, R&D Division, Daiichi Sankyo Co. Ltd, 1-2-58 Hiromachi, Shinagawa-ku, Tokyo 140-8710, Japan. ichikawa.masanori.uf@daiichisankyo.co.jp