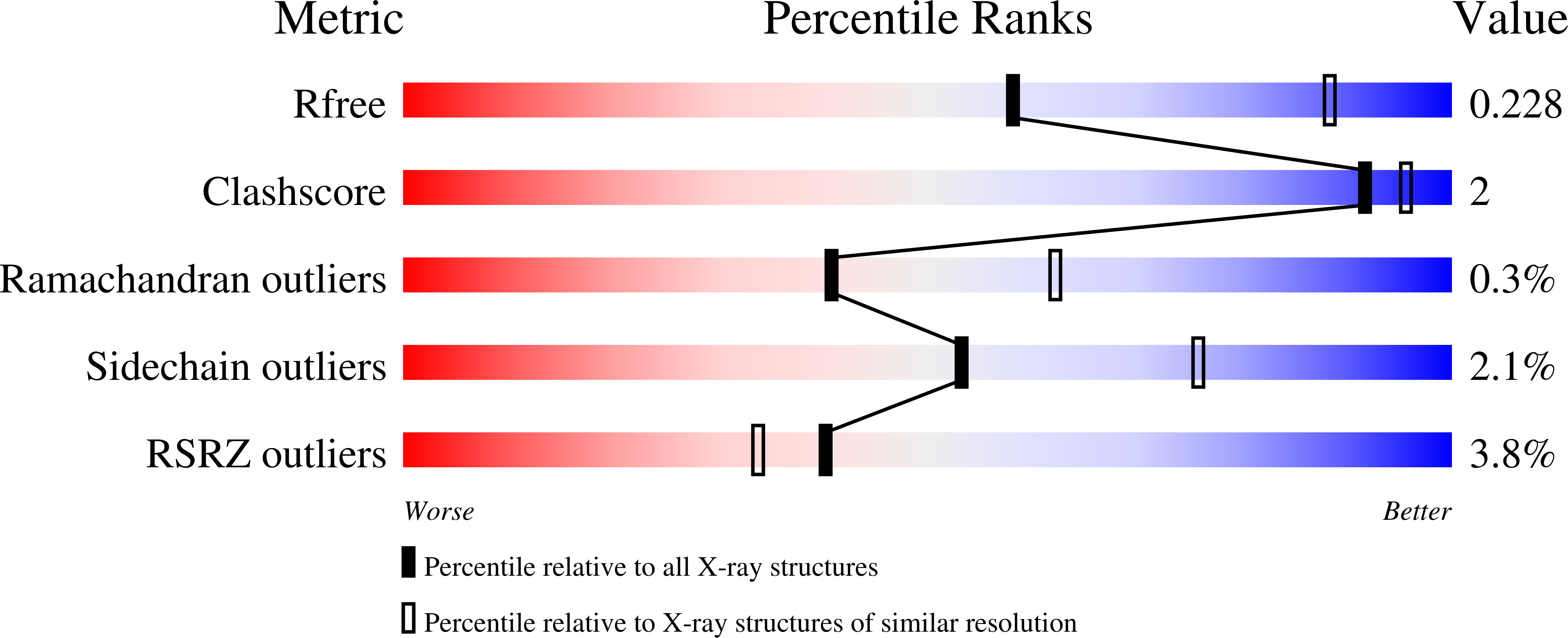
Cation-selective pathway of OmpF porin revealed by anomalous X-ray diffraction.
Dhakshnamoorthy, B., Raychaudhury, S., Blachowicz, L., Roux, B.(2010) J Mol Biol 396: 293-300
- PubMed: 19932117
- DOI: https://doi.org/10.1016/j.jmb.2009.11.042
- Primary Citation of Related Structures:
3HW9, 3HWB - PubMed Abstract:
The OmpF porin from the Escherichia coli outer membrane folds into a trimer of beta-barrels, each forming a wide aqueous pore allowing the passage of ions and small solutes. A long loop (L3) carrying multiple acidic residues folds into the beta-barrel pore to form a narrow "constriction zone". A strong and highly conserved charge asymmetry is observed at the constriction zone, with multiple basic residues attached to the wall of the beta-barrel (Lys16, Arg42, Arg82 and Arg132) on one side, and multiple acidic residues of L3 (Asp107, Asp113, Glu117, Asp121, Asp126, Asp127) on the other side. Several computational studies have suggested that a strong transverse electric field could exist at the constriction zone as a result of such charge asymmetry, giving rise to separate permeation pathways for cations and anions. To examine this question, OmpF was expressed, purified and crystallized in the P6(3) space group and two different data sets were obtained at 2.6 A and 3.0 A resolution with K(+) and Rb(+), respectively. The Rb(+)-soaked crystals were collected at the rubidium anomalous wavelength of 0.8149 A and cation positions were determined. A PEG molecule was observed in the pore region for both the K(+) and Rb(+)-soaked crystals, where it interacts with loop L3. The results reveal the separate pathways of anions and cations across the constriction zone of the OmpF pore.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Gordon Center for Integrative Science, University of Chicago, Chicago, IL 60637, USA.