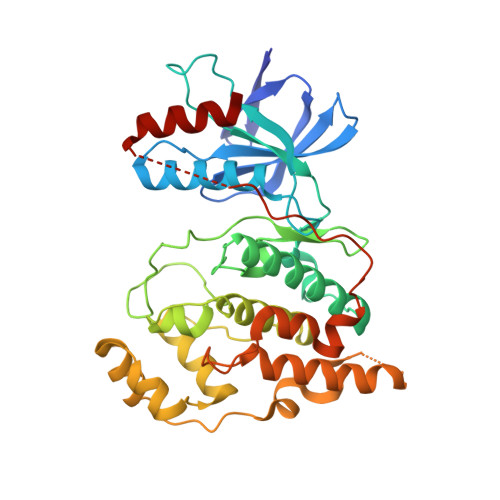
The crystal structure of JNK2 reveals conformational flexibility in the MAP kinase insert and indicates its involvement in the regulation of catalytic activity.
Shaw, D., Wang, S.M., Villasenor, A.G., Tsing, S., Walter, D., Browner, M.F., Barnett, J., Kuglstatter, A.(2008) J Mol Biol 383: 885-893
- PubMed: 18801372
- DOI: https://doi.org/10.1016/j.jmb.2008.08.086
- Primary Citation of Related Structures:
3E7O - PubMed Abstract:
c-Jun N-terminal kinase (JNK) 2 is a member of the mitogen-activated protein (MAP) kinase group of signaling proteins. MAP kinases share a common sequence insertion called "MAP kinase insert", which, for ERK2, has been shown to interact with regulatory proteins and, for p38alpha, has been proposed to be involved in the regulation of catalytic activity. We have determined the crystal structure of human JNK2 complexed with an indazole inhibitor by applying a high-throughput protein engineering and surface-site mutagenesis approach. A novel conformation of the activation loop is observed, which is not compatible with its phosphorylation by upstream kinases. This activation inhibitory conformation of JNK2 is stabilized by the MAP kinase insert that interacts with the activation loop in an induced-fit manner. We therefore suggest that the MAP kinase insert of JNK2 plays a role in the regulation of JNK2 activation, possibly by interacting with intracellular binding partners.
Organizational Affiliation:
Discovery Technologies, Roche Palo Alto, 3431 Hillview Avenue, Palo Alto, CA 94304, USA. david.shaw@roche.com