Crystal Structure of a Putative Transcarbamoylase from Enterococcus Faecalis with Carbamoyl Phosphate
Polo, L.M., Rubio, V.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ASPARTATE/ORNITHINE CARBAMOYLTRANSFERASE | 418 | Enterococcus faecalis | Mutation(s): 0 EC: 2.1.3 | 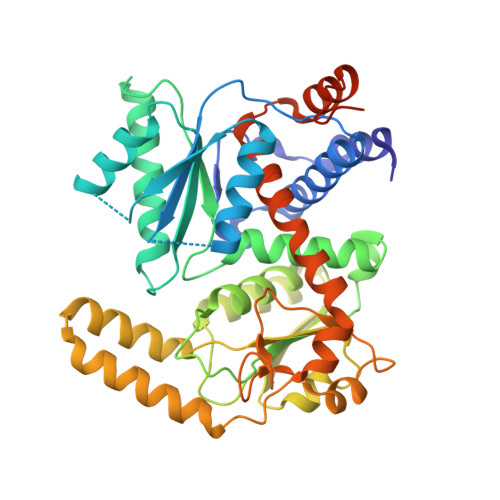 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 117.368 | α = 90 |
| b = 117.368 | β = 90 |
| c = 120.232 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |
| MOLREP | phasing |