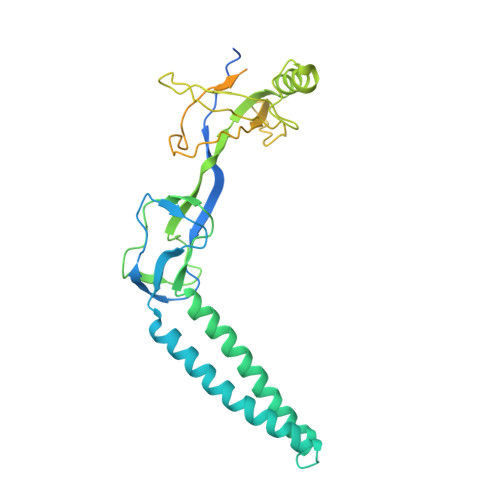
The Assembled Structure of a Complete Tripartite Bacterial Multidrug Efflux Pump.
Symmons, M.F., Bokma, E., Koronakis, E., Hughes, C., Koronakis, V.(2009) Proc Natl Acad Sci U S A 106: 7173
- PubMed: 19342493
- DOI: https://doi.org/10.1073/pnas.0900693106
- Primary Citation of Related Structures:
2V4D - PubMed Abstract:
Bacteria like Escherichia coli and Pseudomonas aeruginosa expel drugs via tripartite multidrug efflux pumps spanning both inner and outer membranes and the intervening periplasm. In these pumps a periplasmic adaptor protein connects a substrate-binding inner membrane transporter to an outer membrane-anchored TolC-type exit duct. High-resolution structures of all 3 components are available, but a pump model has been precluded by the incomplete adaptor structure, because of the apparent disorder of its N and C termini. We reveal that the adaptor termini assemble a beta-roll structure forming the final domain adjacent to the inner membrane. The completed structure enabled in vivo cross-linking to map intermolecular contacts between the adaptor AcrA and the transporter AcrB, defining a periplasmic interface between several transporter subdomains and the contiguous beta-roll, beta-barrel, and lipoyl domains of the adaptor. With short and long cross-links expressed as distance restraints, the flexible linear topology of the adaptor allowed a multidomain docking approach to model the transporter-adaptor complex, revealing that the adaptor docks to a transporter region of comparative stability distinct from those key to the proposed rotatory pump mechanism, putative drug-binding pockets, and the binding site of inhibitory DARPins. Finally, we combined this docking with our previous resolution of the AcrA hairpin-TolC interaction to develop a model of the assembled tripartite complex, satisfying all of the experimentally-derived distance constraints. This AcrA(3)-AcrB(3)-TolC(3) model presents a 610,000-Da, 270-A-long efflux pump crossing the entire bacterial cell envelope.
Organizational Affiliation:
Department of Pathology, Cambridge University, Tennis Court Road, Cambridge CB2 1QP, United Kingdom.