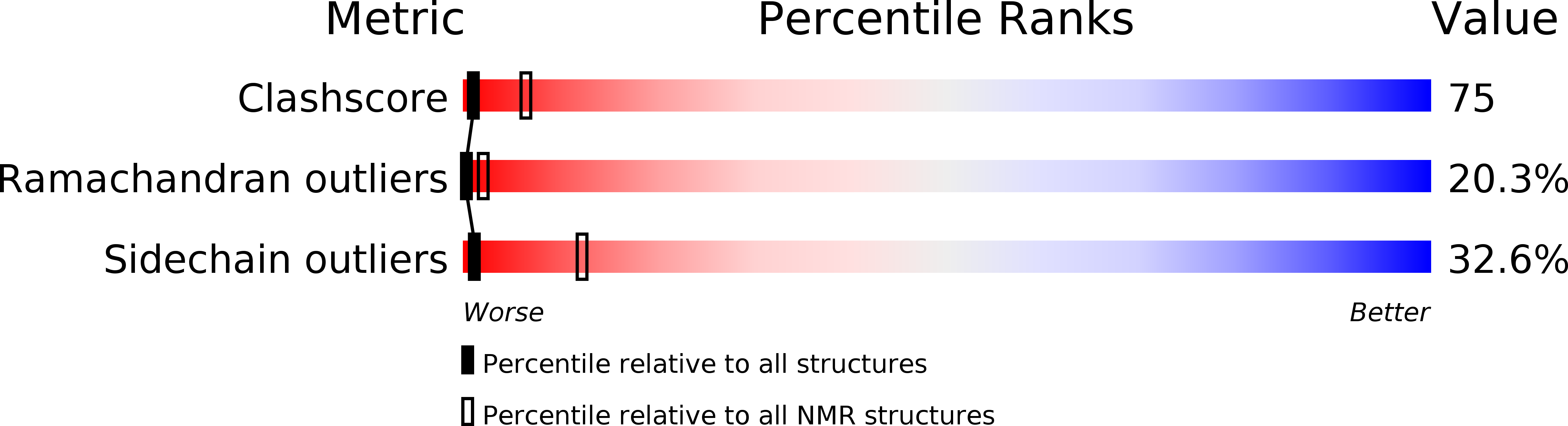Solution structure of R-elafin, a specific inhibitor of elastase.
Francart, C., Dauchez, M., Alix, A.J., Lippens, G.(1997) J Mol Biol 268: 666-677
- PubMed: 9171290
- DOI: https://doi.org/10.1006/jmbi.1997.0983
- Primary Citation of Related Structures:
2REL - PubMed Abstract:
The solution structure of r-elafin, a specific elastase inhibitor, has been determined using NMR spectroscopy. Characterized by a flat core and a flexible N-terminal extremity, the three-dimensional structure is formed by a central twisted beta-hairpin accompanied by two external segments linked by the proteinase binding loop. A cluster of three disulfide bridges connects the external segments to the central beta-sheet and a single fourth disulfide bridge links the binding loop to the central beta-turn. The same spatial distribution of disulfide bridges can be observed in both domains of the secretory leukocyte protease inhibitor (SLPI), another elastase inhibitor. The structural homology between r-elafin and the C-terminal domain of SLPI confirms the former as a second member of the chelonianin family of proteinase inhibitors. Based on the homology between the two proteins and recent results obtained for elastase binding mutants of the bovine pancreatic trypsin inhibitor (BPTI), we define the segment 22 to 27 as the binding loop of elafin, with the scissile peptide bond between Ala24 and Met25. In our solution structures, this loop is extended and solvent-exposed, and exhibits a large degree of flexibility. This mobility, already observed for the binding loop in other protease inhibitors in solution, might be an important feature for the interaction with the corresponding protease.
Organizational Affiliation:
CNRS URA 1309, Institut Pasteur de Lille, France.