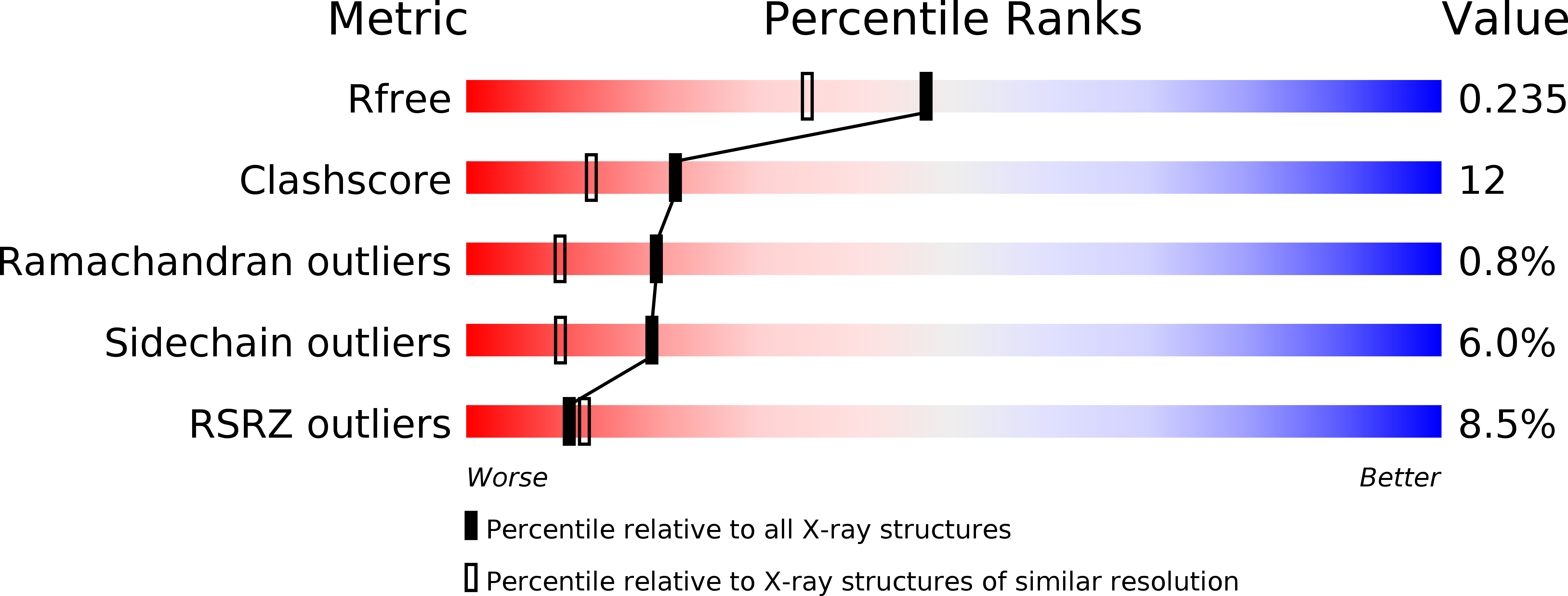
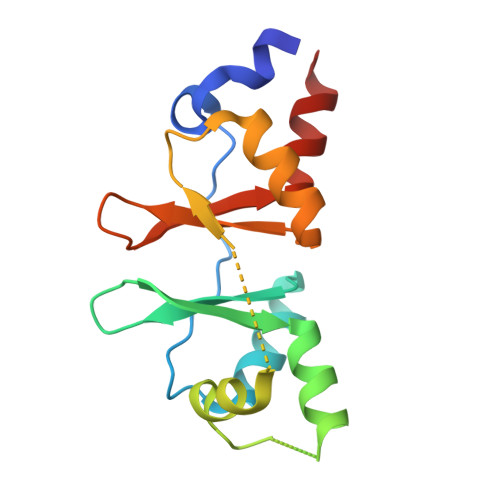
Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding.
Rudolph, M.J., Amodeo, G.A., Iram, S., Hong, S.P., Pirino, G., Carlson, M., Tong, L.(2007) Structure 15: 65-74
- PubMed: 17223533
- DOI: https://doi.org/10.1016/j.str.2006.11.014
- Primary Citation of Related Structures:
2NYC, 2NYE - PubMed Abstract:
AMP-activated protein kinase (AMPK) is a central regulator of energy homeostasis in mammals. AMP is believed to control the activity of AMPK by binding to the gamma subunit of this heterotrimeric enzyme. This subunit contains two Bateman domains, each of which is composed of a tandem pair of cystathionine beta-synthase (CBS) motifs. No structural information is currently available on this subunit, and the molecular basis for its interactions with AMP is not well understood. We report here the crystal structure at 1.9 Angstrom resolution of the Bateman2 domain of Snf4, the gamma subunit of the yeast ortholog of AMPK. The structure revealed a dimer of the Bateman2 domain, and this dimerization is supported by our light-scattering, mutagenesis, and biochemical studies. There is a prominent pocket at the center of this dimer, and most of the disease-causing mutations are located in or near this pocket.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, NY 10027, USA.