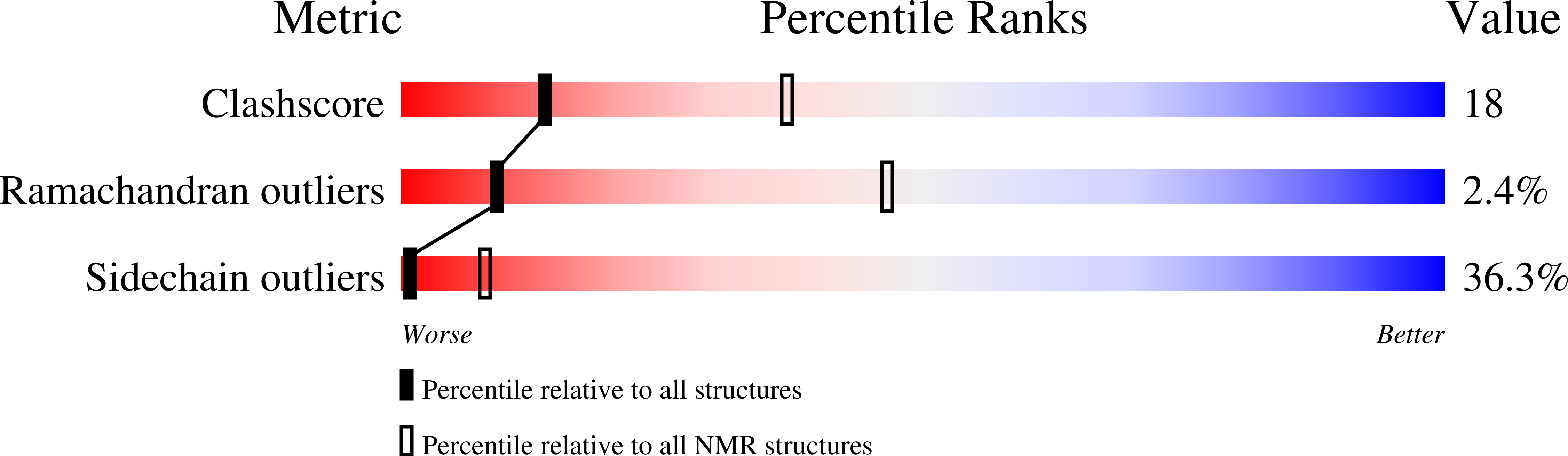Antiparallel coiled-coil-mediated dimerization of myosin X
Lu, Q., Ye, F., Wei, Z., Wen, Z., Zhang, M.(2012) Proc Natl Acad Sci U S A 109: 17388-17393
- PubMed: 23012428
- DOI: https://doi.org/10.1073/pnas.1208642109
- Primary Citation of Related Structures:
2LW9 - PubMed Abstract:
Processive movements of unconventional myosins on actin filaments generally require motor dimerization. A commonly accepted myosin dimerization mechanism is via formation of a parallel coiled-coil dimer by a stretch of amino acid residues immediately carboxyl-terminal to the motor's lever-arm domain. Here, we discover that the predicted coiled-coil region of myosin X forms a highly stable, antiparallel coiled-coil dimer (anti-CC). Disruption of the anti-CC either by single-point mutations or by replacement of the anti-CC with a parallel coiled coil with a similar length compromised the filopodial induction activity of myosin X. We further show that the anti-CC and the single α-helical domain of myosin X are connected by a semirigid helical linker. The anti-CC-mediated dimerization may enable myosin X to walk on both single and bundled actin filaments.
Organizational Affiliation:
Division of Life Science, State Key Laboratory of Molecular Neuroscience and Center of Systems Biology and Human Health, School of Science and Institute for Advanced Study, Hong Kong University of Science and Technology, Kowloon, Hong Kong, China.