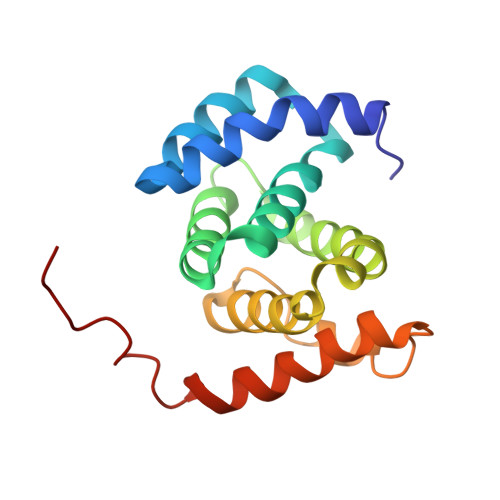
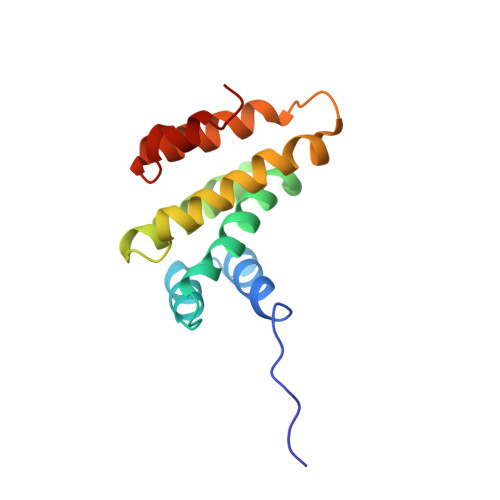
Structure of the tandem MA-3 region of Pdcd4 protein and characterization of its interactions with eIF4A and eIF4G: molecular mechanisms of a tumor suppressor
Waters, L.C., Strong, S.L., Ferlemann, E., Oka, O., Muskett, F.W., Veverka, V., Banerjee, S., Schmedt, T., Henry, A.J., Klempnauer, K.H., Carr, M.D.(2011) J Biol Chem 286: 17270-17280
- PubMed: 21454508
- DOI: https://doi.org/10.1074/jbc.M110.166157
- Primary Citation of Related Structures:
2KZT - PubMed Abstract:
One of the key regulatory points of translation initiation is recruitment of the 43S preinitation complex to the 5' mRNA cap by the eIF4F complex (eIF4A, eIF4E, and eIF4G). The tumor suppressor protein Pdcd4 has been shown to inhibit cap-dependent translation by interacting tightly with the RNA helicase eIF4A via its tandem MA-3 domains. The NMR studies reported here reveal a fairly extensive and well defined interface between the two MA-3 domains in solution, which appears to be stabilized by a network of interdomain salt bridges and hydrogen bonds, and reveals a unique orientation of the two domains. Characterization of the stoichiometry of the Pdcd4-eIF4A complex suggests that under physiological conditions Pdcd4 binds to a single molecule of eIF4A, which involves contacts with both Pdcd4 MA-3 domains. We also show that contacts mediated by a conserved acidic patch on the middle MA-3 domain of Pdcd4 are essential for forming a tight complex with eIF4A in vivo, whereas the equivalent region of the C-terminal MA-3 domain appears to have no role in complex formation in vivo. The formation of a 1:1 eIF4A-Pdcd4 complex in solution is consistent with the reported presence in vivo of only one molecule of eIF4A in the eIF4F complex. Pdcd4 has also been reported to interact directly with the middle region of eIF4G, however, we were unable to obtain any evidence for even a weak, transient direct interaction.
Organizational Affiliation:
Department of Biochemistry, Henry Wellcome Building, University of Leicester, Lancaster Road, Leicester LE1 9HN, United Kingdom. lw83@le.ac.uk