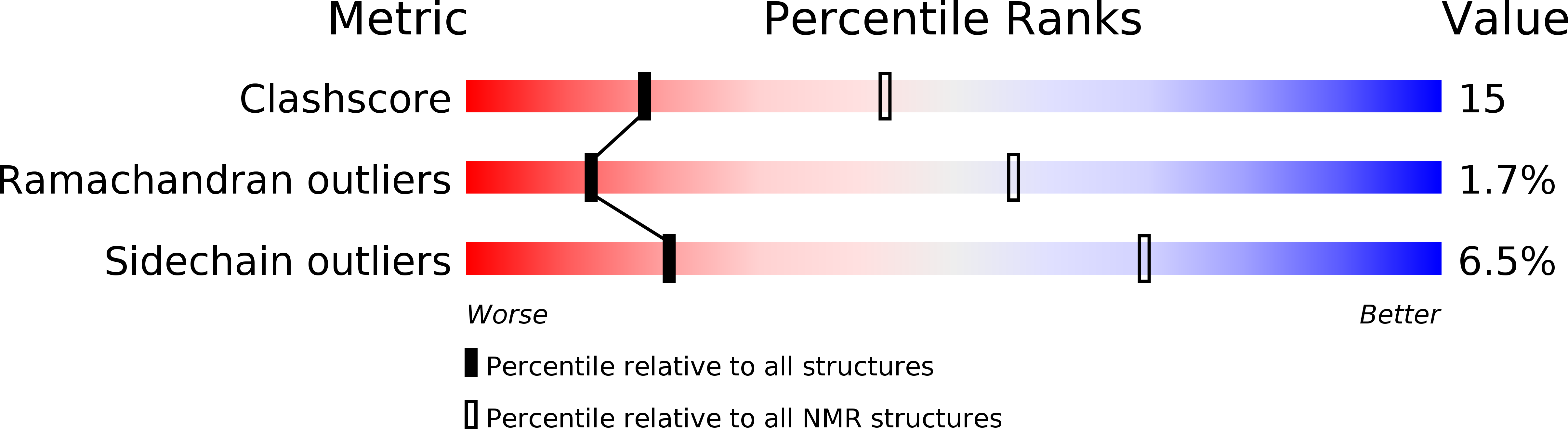NMR solution structure of SlyD from Escherichia coli: spatial separation of prolyl isomerase and chaperone function.
Weininger, U., Haupt, C., Schweimer, K., Graubner, W., Kovermann, M., Bruser, T., Scholz, C., Schaarschmidt, P., Zoldak, G., Schmid, F.X., Balbach, J.(2009) J Mol Biol 387: 295-305
- PubMed: 19356587
- DOI: https://doi.org/10.1016/j.jmb.2009.01.034
- Primary Citation of Related Structures:
2K8I - PubMed Abstract:
SlyD (sensitive to lysis D) is a putative folding helper from the bacterial cytosol and harbors prolyl isomerase and chaperone activities. We determined the solution NMR structure of a truncated version of SlyD (1-165) from Escherichia coli (SlyD*) that lacks the presumably unstructured C-terminal tail. SlyD* consists of two well-separated domains: the FKBP domain, which harbors the prolyl isomerase activity, and the insert-in-flap (IF) domain, which harbors the chaperone activity. The IF domain is inserted into a loop of the FKBP domain near the prolyl isomerase active site. The NMR structure of SlyD* showed no distinct orientation of the two domains relative to each other. In the FKBP domain, Tyr68 points into the active site, which might explain the lowered intrinsic prolyl isomerase activity and the much lower FK506 binding affinity of the protein compared with archetype human FKBP12 (human FK506 binding protein with 12 kDa). The thermodynamics and kinetics of substrate binding by SlyD* were quantified by fluorescence resonance energy transfer. NMR titration experiments revealed that the IF domain recognizes and binds unfolded or partially folded proteins and peptides. Insulin aggregation is markedly slowed by SlyD* as evidenced by two-dimensional NMR spectroscopy in real time, probably due to SlyD* binding to denatured insulin. The capacity of the IF domain to establish an initial encounter-collision complex, together with the flexible orientation of the two interacting domains, makes SlyD* a very powerful catalyst of protein folding.
Organizational Affiliation:
Institut für Physik, Fachgruppe Biophysik, Martin-Luther-Universität Halle-Wittenberg, D-06099 Halle (Saale), Germany.