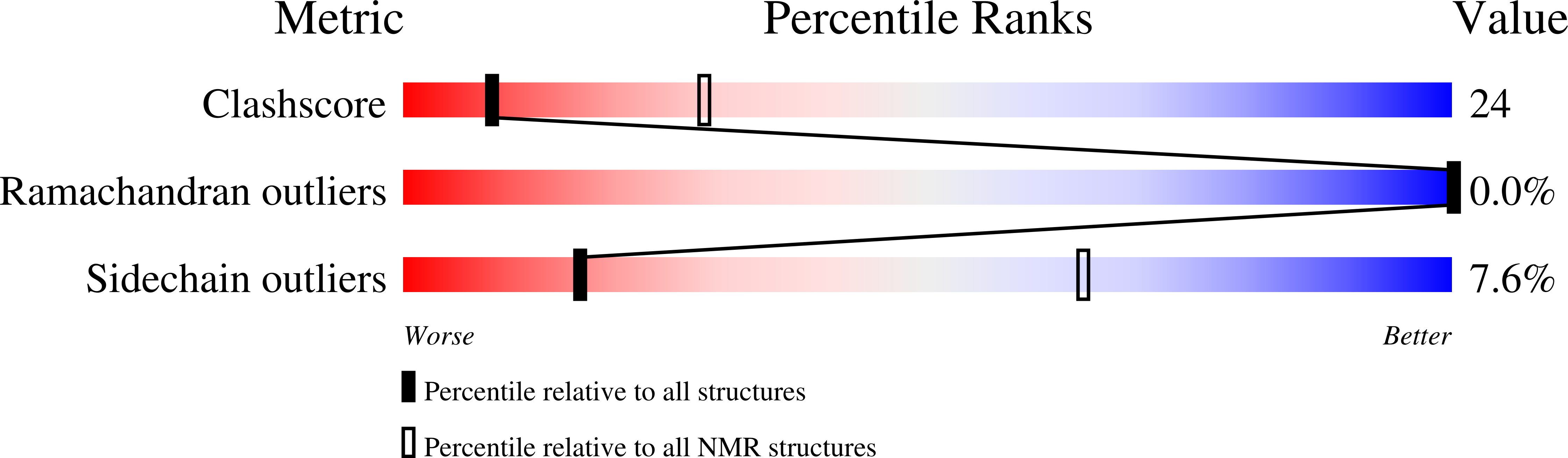Doublesex and the Regulation of Sexual Dimorphism in Drosophila melanogaster: STRUCTURE, FUNCTION, AND MUTAGENESIS OF A FEMALE-SPECIFIC DOMAIN.
Yang, Y., Zhang, W., Bayrer, J.R., Weiss, M.A.(2008) J Biol Chem 283: 7280-7292
- PubMed: 18184648
- DOI: https://doi.org/10.1074/jbc.M708742200
- Primary Citation of Related Structures:
2JZ0, 2JZ1 - PubMed Abstract:
The DSX (Doublesex) transcription factor regulates somatic sexual differentiation in Drosophila. Female and male isoforms (DSX F and DSX M) are formed due to sex-specific RNA splicing. DNA recognition, mediated by a shared N-terminal zinc module (the DM domain), is enhanced by a C-terminal dimerization element. Sex-specific extension of this element in DSX F and DSX M leads to assembly of distinct transcriptional preinitiation complexes. Here, we describe the structure of the extended C-terminal dimerization domain of DSX F as determined by multidimensional NMR spectroscopy. The core dimerization element is well ordered, giving rise to a dense network of interresidue nuclear Overhauser enhancements. The structure contains dimer-related UBA folds similar to those defined by x-ray crystallographic studies of a truncated domain. Whereas the proximal portion of the female tail extends helix 3 of the UBA fold, the distal tail is disordered. Ala substitutions in the proximal tail disrupt the sex-specific binding of IX (Intersex), an obligatory partner protein and putative transcriptional coactivator; IX-DSX F interaction is, by contrast, not disrupted by truncation of the distal tail. Mutagenesis of the UBA-like dimer of DSX F highlights the importance of steric and electrostatic complementarity across the interface. Two temperature-sensitive mutations at this interface have been characterized in yeast model systems. One weakens a network of solvated salt bridges, whereas the other perturbs the underlying nonpolar interface. These mutations confer graded gene-regulatory activity in yeast within a physiological temperature range and so may provide novel probes for genetic analysis of a sex-specific transcriptional program in Drosophila development.
Organizational Affiliation:
Department of Biochemistry, Case Western Reserve University, Cleveland, Ohio 44106, USA.