Distinct functional domains of ubc9 dictate cell survival and resistance to genotoxic stress.
van Waardenburg, R.C., Duda, D.M., Lancaster, C.S., Schulman, B.A., Bjornsti, M.A.(2006) Mol Cell Biol 26: 4958-4969
- PubMed: 16782883
- DOI: https://doi.org/10.1128/MCB.00160-06
- Primary Citation of Related Structures:
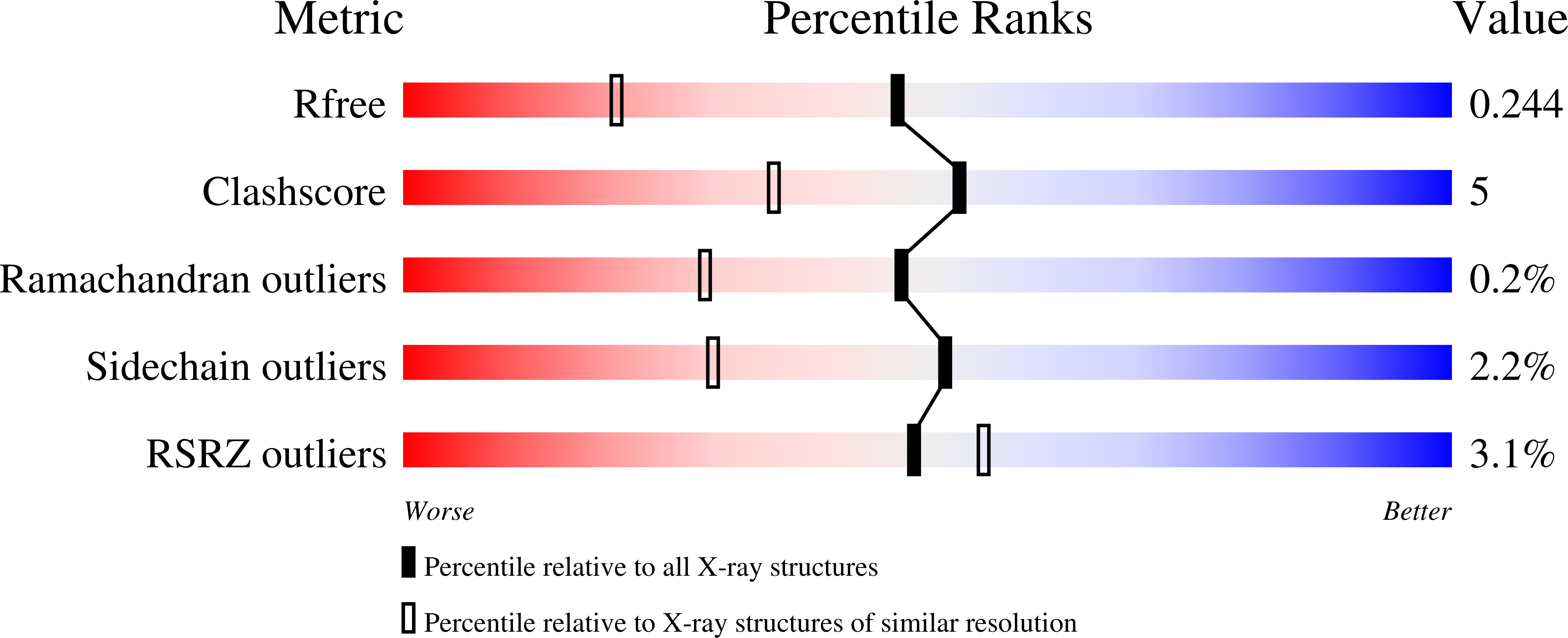
2GJD - PubMed Abstract:
Covalent modification with SUMO alters protein function, intracellular localization, or protein-protein interactions. Target recognition is determined, in part, by the SUMO E2 enzyme, Ubc9, while Siz/Pias E3 ligases may facilitate select interactions by acting as substrate adaptors. A yeast conditional Ubc9P(123)L mutant was viable at 36 degrees C yet exhibited enhanced sensitivity to DNA damage. To define functional domains in Ubc9 that dictate cellular responses to genotoxic stress versus those necessary for cell viability, a 1.75-A structure of yeast Ubc9 that demonstrated considerable conservation of backbone architecture with human Ubc9 was solved. Nevertheless, differences in side chain geometry/charge guided the design of human/yeast chimeras, where swapping domains implicated in (i) binding residues within substrates that flank canonical SUMOylation sites, (ii) interactions with the RanBP2 E3 ligase, and (iii) binding of the heterodimeric E1 and SUMO had distinct effects on cell growth and resistance to DNA-damaging agents. Our findings establish a functional interaction between N-terminal and substrate-binding domains of Ubc9 and distinguish the activities of E3 ligases Siz1 and Siz2 in regulating cellular responses to genotoxic stress.
Organizational Affiliation:
Dept. of Molecular Pharmacology, St. Jude Children's Research Hospital, 332 N. Lauderdale, Memphis, TN 38105, USA.