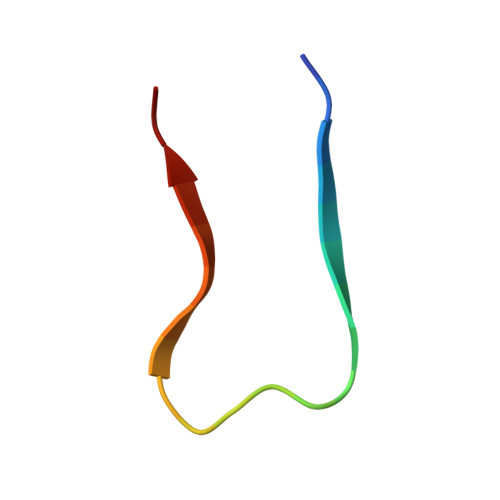
3D structure of amyloid protofilaments of beta2-microglobulin fragment probed by solid-state NMR
Iwata, K., Fujiwara, T., Matsuki, Y., Akutsu, H., Takahashi, S., Naiki, H., Goto, Y.(2006) Proc Natl Acad Sci U S A 103: 18119-18124
- PubMed: 17108084
- DOI: https://doi.org/10.1073/pnas.0607180103
- Primary Citation of Related Structures:
2E8D - PubMed Abstract:
Understanding the structure and formation of amyloid fibrils, the filamentous aggregates of proteins and peptides, is crucial in preventing diseases caused by their deposition and, moreover, for obtaining further insight into the mechanism of protein folding and misfolding. We have combined solid-state NMR, x-ray fiber diffraction, and atomic force microscopy to reveal the 3D structure of amyloid protofilament-like fibrils formed by a 22-residue K3 peptide (Ser(20)-Lys(41)) of beta(2)-microglobulin, a protein responsible for dialysis-related amyloidosis. Although a uniformly (13)C,(15)N-labeled sample was used for the NMR measurements, we could obtain the 3D structure of the fibrils on the basis of a large number of structural constraints. The conformation of K3 fibrils was found to be a beta-strand-loop-beta-strand with each K3 molecule stacked in a parallel and staggered manner. It is suggested that the fibrillar conformation is stabilized by intermolecular interactions, rather than by intramolecular hydrophobic packing as seen in globular proteins. Together with thermodynamic studies of the full-length protein, formation of the fibrils is likely to require side chains on the intermolecular surface to pack tightly against those of adjacent monomers. By revealing the structure of beta(2)-microglobulin protofilament-like fibrils, this work represents technical progress in analyzing amyloid fibrils in general through solid-state NMR.
Organizational Affiliation:
Institute for Protein Research, Osaka University and Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, Suita, Osaka 565-0871, Japan.