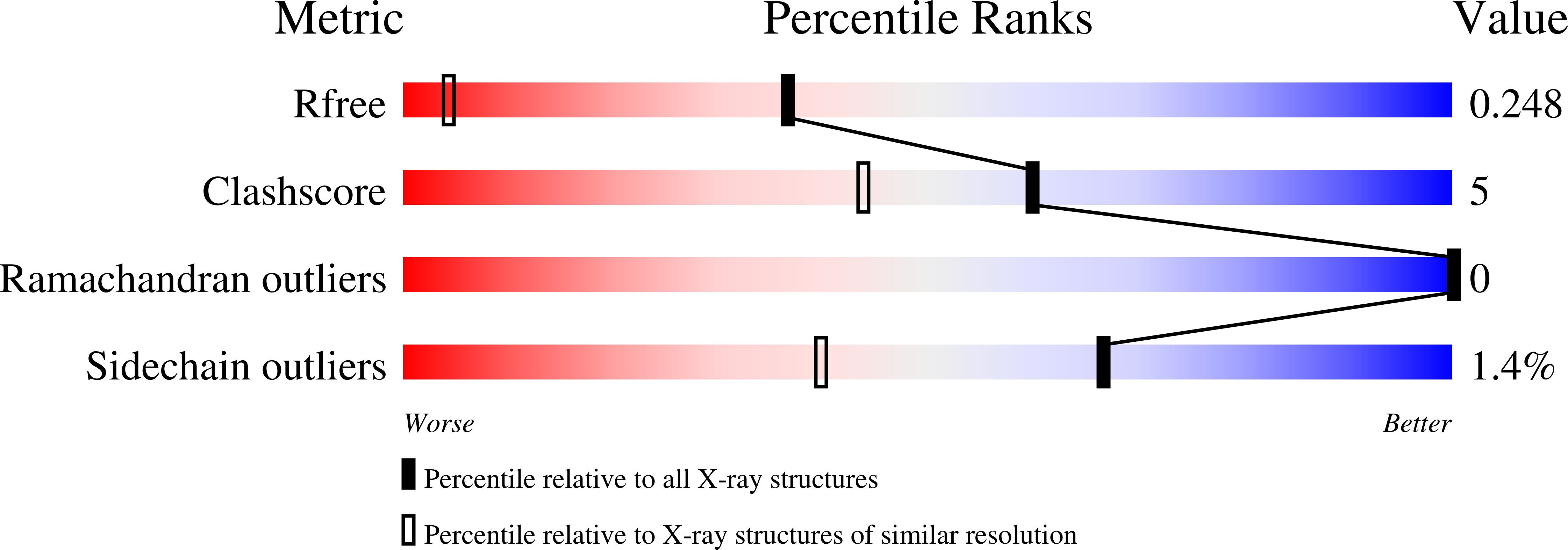
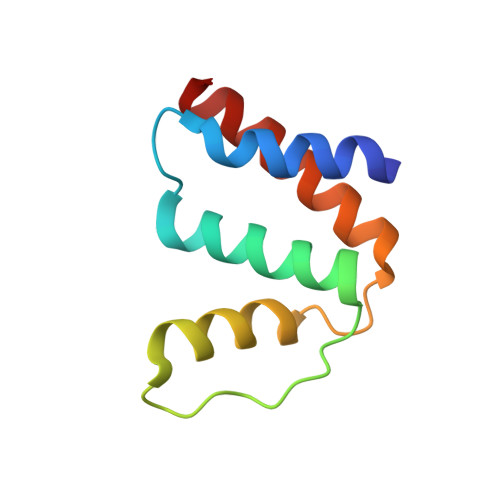
High Resolution Crystal Structures of Unliganded and Liganded Human Liver Acbp Reveal a New Mode of Binding for the Acyl-Coa Ligand.
Taskinen, J.P., Van Aalten, D.M., Knudsen, J., Wierenga, R.K.(2007) Proteins 66: 229
- PubMed: 17044054
- DOI: https://doi.org/10.1002/prot.21124
- Primary Citation of Related Structures:
2CB8, 2FJ9 - PubMed Abstract:
The acyl-CoA binding protein (ACBP) is essential for the fatty acid metabolism, membrane structure, membrane fusion, and ceramide synthesis. Here high resolution crystal structures of human cytosolic liver ACBP, unliganded and liganded with a physiological ligand, myristoyl-CoA are described. The binding of the acyl-CoA molecule induces only few structural differences near the binding pocket. The crystal form of the liganded ACBP, which has two ACBP molecules in the asymmetric unit, shows that in human ACBP the same acyl-CoA binding pocket is present as previously described for the bovine and Plasmodium falciparum ACBP and the mode of binding of the 3'-phosphate-AMP moiety is conserved. Unexpectedly, in one of the acyl-CoA binding pockets the acyl moiety is bound in a reversed mode as compared with the bovine and P. falciparum structures. In this binding mode, the myristoyl-CoA molecule is fully ordered and bound across the two ACBP molecules of the crystallographic asymmetric unit: the 3'-phosphate-AMP moiety is bound in the binding pocket of one ACBP molecule and the acyl chain is bound in the pocket of the other ACBP molecule. The remaining binding pocket cavities of these two ACBP molecules are filled by other ligand fragments. This novel binding mode shows that the acyl moiety can flip out of its classical binding pocket and bind elsewhere, suggesting a mechanism for the acyl-CoA transfer between ACBP and the active site of a target enzyme. This mechanism is of possible relevance for the in vivo function of ACBP.
Organizational Affiliation:
Biocenter Oulu and Department of Biochemistry, University of Oulu, FIN-90014, Finland.