Intrinsic disorder drives N-terminal ubiquitination by Ube2w.
Vittal, V., Shi, L., Wenzel, D.M., Scaglione, K.M., Duncan, E.D., Basrur, V., Elenitoba-Johnson, K.S., Baker, D., Paulson, H.L., Brzovic, P.S., Klevit, R.E.(2015) Nat Chem Biol 11: 83-89
- PubMed: 25436519
- DOI: https://doi.org/10.1038/nchembio.1700
- Primary Citation of Related Structures:
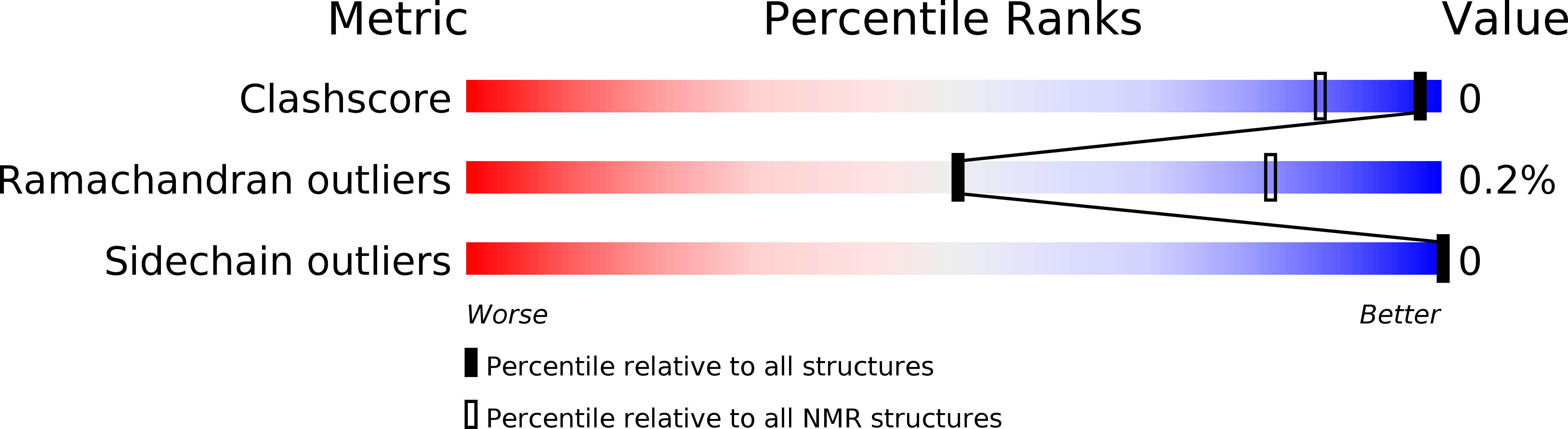
2MT6 - PubMed Abstract:
Ubiquitination of the αN-terminus of protein substrates has been reported sporadically since the early 1980s. However, the identity of an enzyme responsible for this unique ubiquitin (Ub) modification has only recently been elucidated. We show the Ub-conjugating enzyme (E2) Ube2w uses a unique mechanism to facilitate the specific ubiquitination of the α-amino group of its substrates that involves recognition of backbone atoms of intrinsically disordered N termini. We present the NMR-based solution ensemble of full-length Ube2w that reveals a structural architecture unlike that of any other E2 in which its C terminus is partly disordered and flexible to accommodate variable substrate N termini. Flexibility of the substrate is critical for recognition by Ube2w, and either point mutations in or the removal of the flexible C terminus of Ube2w inhibits substrate binding and modification. Mechanistic insights reported here provide guiding principles for future efforts to define the N-terminal ubiquitome in cells.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, Washington, USA.