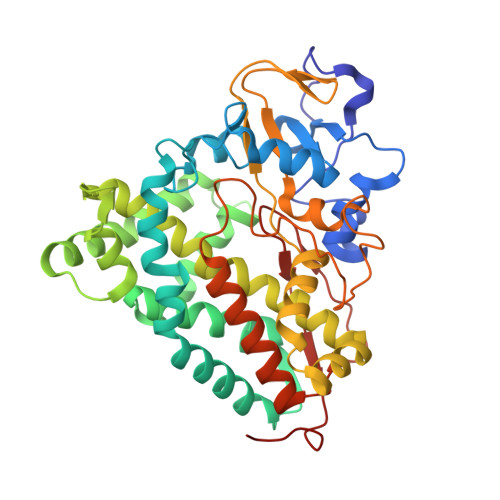
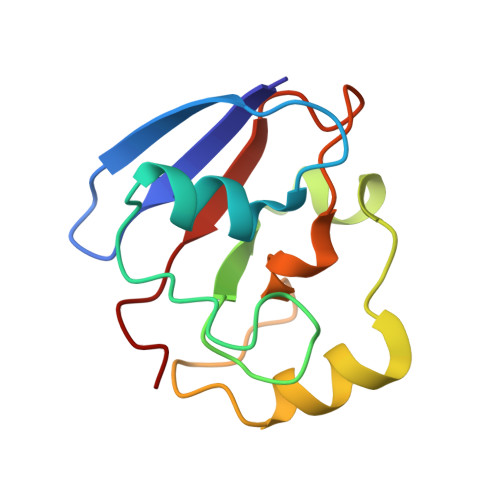
The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography.
Hiruma, Y., Hass, M.A., Kikui, Y., Liu, W.M., Olmez, B., Skinner, S.P., Blok, A., Kloosterman, A., Koteishi, H., Lohr, F., Schwalbe, H., Nojiri, M., Ubbink, M.(2013) J Mol Biol 425: 4353-4365
- PubMed: 23856620
- DOI: https://doi.org/10.1016/j.jmb.2013.07.006
- Primary Citation of Related Structures:
2M56, 3W9C - PubMed Abstract:
Cytochrome P450cam catalyzes the hydroxylation of camphor in a complex process involving two electron transfers (ETs) from the iron-sulfur protein putidaredoxin. The enzymatic control of the successive steps of catalysis is critical for a highly efficient reaction. The injection of the successive electrons is part of the control system. To understand the molecular interactions between putidaredoxin and cytochrome P450cam, we determined the structure of the complex both in solution and in the crystal state. Paramagnetic NMR spectroscopy using lanthanide tags yielded 446 structural restraints that were used to determine the solution structure. An ensemble of 10 structures with an RMSD of 1.3Å was obtained. The crystal structure of the complex was solved, showing a position of putidaredoxin that is identical with the one in the solution structure. The NMR data further demonstrate the presence of a minor state or set of states of the complex in solution, which is attributed to the presence of an encounter complex. The structure of the major state shows a small binding interface and a metal-to-metal distance of 16Å, with two pathways that provide strong electronic coupling of the redox centers. The interpretation of these results is discussed in the context of ET. The structure indicates that the ET rate can be much faster than the reported value, suggesting that the process may be gated.
Organizational Affiliation:
Leiden Institute of Chemistry, Gorlaeus Laboratories, Leiden University, Einsteinweg 55, 2333 CC Leiden, The Netherlands. Electronic address: hirumay@chem.leidenuniv.nl.