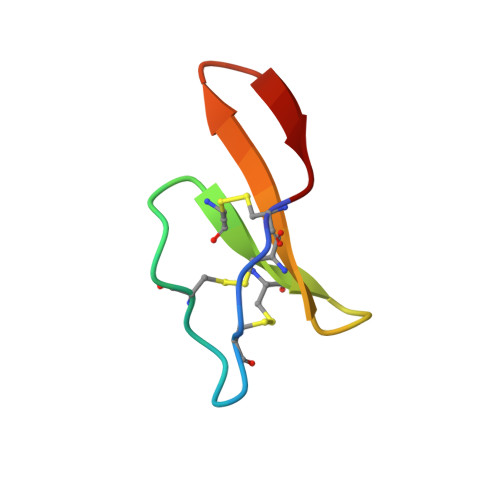
Processing of a 22 kDa precursor protein to produce the circular protein tricyclon A.
Mulvenna, J.P., Sando, L., Craik, D.J.(2005) Structure 13: 691-701
- PubMed: 15893660
- DOI: https://doi.org/10.1016/j.str.2005.02.013
- Primary Citation of Related Structures:
1YP8 - PubMed Abstract:
Cyclotides are a family of plant proteins that have the unusual combination of head-to-tail backbone cyclization and a cystine knot motif. They are exceptionally stable and show resistance to most chemical, physical, and enzymatic treatments. The structure of tricyclon A, a previously unreported cyclotide, is described here. In this structure, a loop that is disordered in other cyclotides forms a beta sheet that protrudes from the globular core. This study indicates that the cyclotide fold is amenable to the introduction of a range of structural elements without affecting the cystine knot core of the protein, which is essential for the stability of the cyclotides. Tricyclon A does not possess a hydrophobic patch, typical of other cyclotides, and has minimal hemolytic activity, making it suitable for pharmaceutical applications. The 22 kDa precursor protein of tricyclon A was identified and provides clues to the processing of these fascinating miniproteins.
Organizational Affiliation:
Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia.