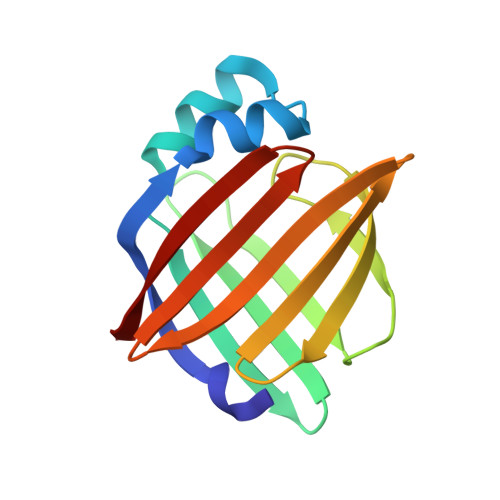
Structure of myelin P2 protein from equine spinal cord.
Hunter, D.J., Macmaster, R., Roszak, A.W., Riboldi-Tunnicliffe, A., Griffiths, I.R., Freer, A.A.(2005) Acta Crystallogr D Biol Crystallogr 61: 1067-1071
- PubMed: 16041071
- DOI: https://doi.org/10.1107/S0907444905014162
- Primary Citation of Related Structures:
1YIV - PubMed Abstract:
Equine P2 protein has been isolated from horse spinal cord and its structure determined to 2.1 A. Since equine myelin is a viable alternative to bovine tissue for large-scale preparations, characterization of the proteins from equine spinal cord myelin has been initiated. There is an unusually high amount of P2 protein in equine CNS myelin compared with other species. The structure was determined by molecular replacement and subsequently refined to an R value of 0.187 (Rfree=0.233). The structure contains a molecule of the detergent LDAO and HEPES buffer in the binding cavity and is otherwise analogous to other cellular retinol-binding proteins.
Organizational Affiliation:
Department of Chemistry, Joseph Black Building, University of Glasgow, Glasgow G12 8QQ, Scotland.