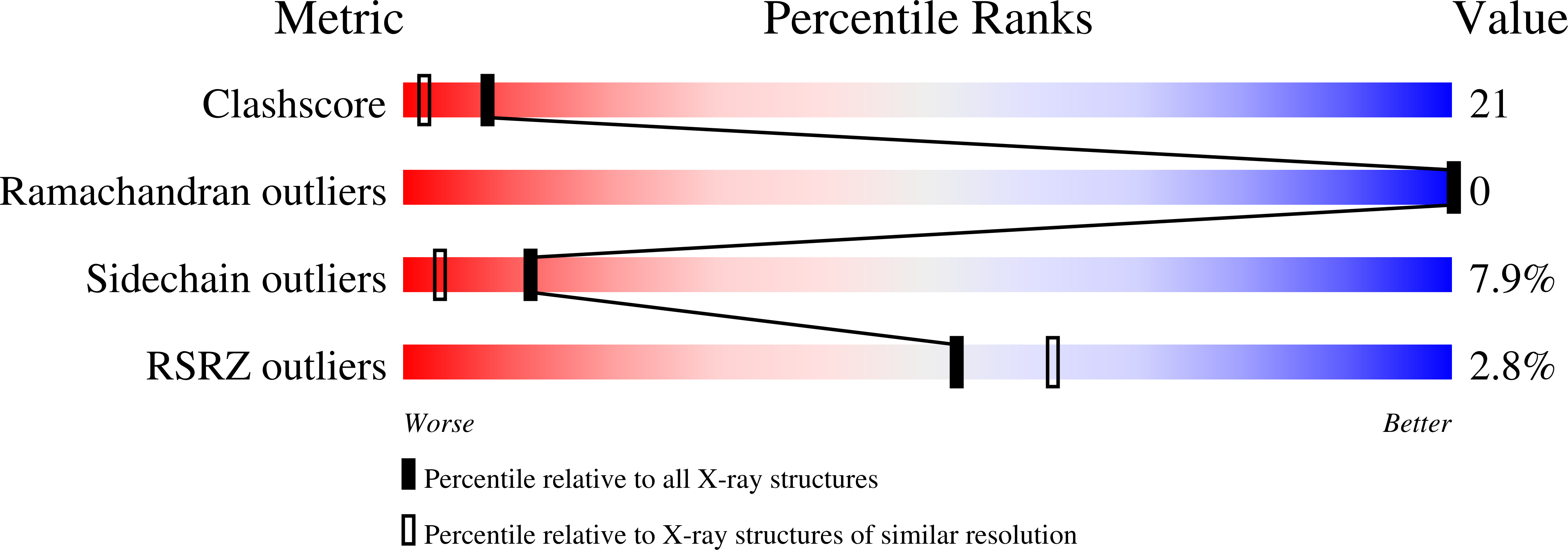Structure determination and analysis of yeast iso-2-cytochrome c and a composite mutant protein.
Murphy, M.E., Nall, B.T., Brayer, G.D.(1992) J Mol Biol 227: 160-176
- PubMed: 1326054
- DOI: https://doi.org/10.1016/0022-2836(92)90689-h
- Primary Citation of Related Structures:
1YEA, 1YEB - PubMed Abstract:
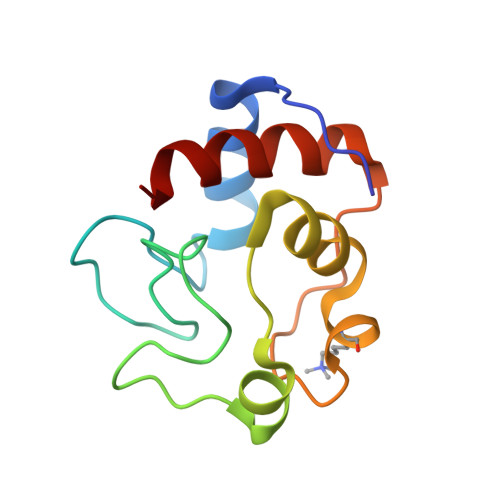
As part of a study of protein folding and stability, the three-dimensional structures of yeast iso-2-cytochrome c and a composite protein (B-2036) composed of primary sequences of both iso-1 and iso-2-cytochromes c have been solved to 1.9 A and 1.95 A resolutions, respectively, using X-ray diffraction techniques. The sequences of iso-1 and iso-2-cytochrome c share approximately 84% identity and the B-2036 composite protein has residues 15 to 63 from iso-2-cytochrome c with the rest being derived form the iso-1 protein. Comparison of these structures reveals that amino acid substitutions result in alterations in the details of intramolecular interactions. Specifically, the substitution Leu98Met results in the filling of an internal cavity present in iso-1-cytochrome c. Further substitutions of Val20Ile and Cys102Ala alter the packing of secondary structure elements in the iso-2 protein. Blending the isozymic amino acid sequences in this latter area results in the expansion of the volume of an internal cavity in the B-2036 structure to relieve a steric clash between Ile20 and Cys102. Modification of hydrogen bonding and protein packing without disrupting the protein fold is illustrated by the His26Asn and Asn63Ser substitutions between iso-1 and iso-2-cytochromes c. Alternatively, a change in main-chain fold is observed at Gly37 apparently due to a remote amino acid substitution. Further structural changes occur at Phe82 and the amino terminus where a four residue extension is present in yeast iso-2-cytochrome c. An additional comparison with all other eukaryotic cytochrome c structures determined to date is presented, along with an analysis of conserved water molecules. Also determined are the midpoint reduction potentials of iso-2 and B-2036 cytochromes c using direct electrochemistry. The values obtained are 286 and 288 mV, respectively, indicating that the amino acid substitutions present have had only a small impact on the heme reduction potential in comparison to iso-1-cytochrome c, which has a reduction potential of 290 mV.
Organizational Affiliation:
Department of Biochemistry, University of British Columbia, Vancouver, Canada.