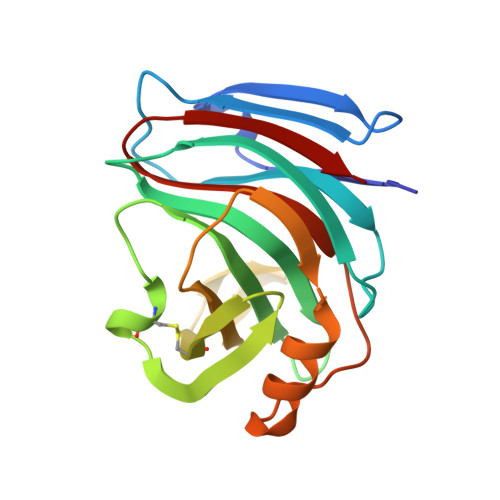
Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum.
Krengel, U., Dijkstra, B.W.(1996) J Mol Biol 263: 70-78
- PubMed: 8890913
- DOI: https://doi.org/10.1006/jmbi.1996.0556
- Primary Citation of Related Structures:
1UKR - PubMed Abstract:
The crystal structure of endo-1,4-beta-xylanase I from Aspergillus niger has been solved by molecular replacement and was refined to 2.4 A resolution. The final R-factor for all data from 6 to 2.4 A is 17.9%. The A. niger xylanase has a characteristic fold which is unique for family G xylanases (root-mean-square deviation = 1.1 A to Trichoderma reesei xylanase I, which has 53% sequence identity). It consists of a single domain composed predominantly of beta-strands. Two beta-sheets are twisted around a deep, long cleft, which is lined with many aromatic amino acid residues and is large enough to accommodate at least four xylose residues. The two conserved glutamate residues, Glu79 and Glu170, which are likely to be involved in catalysis, reach into this cleft from opposite sides. A niger xylanase I is of particular commercial interest because of its low pH optimum. A model is proposed which explains this low pH optimum compared to other members of xylanase family G.
Organizational Affiliation:
BIOSON Research Institute, University of Groningen, The Netherlands.