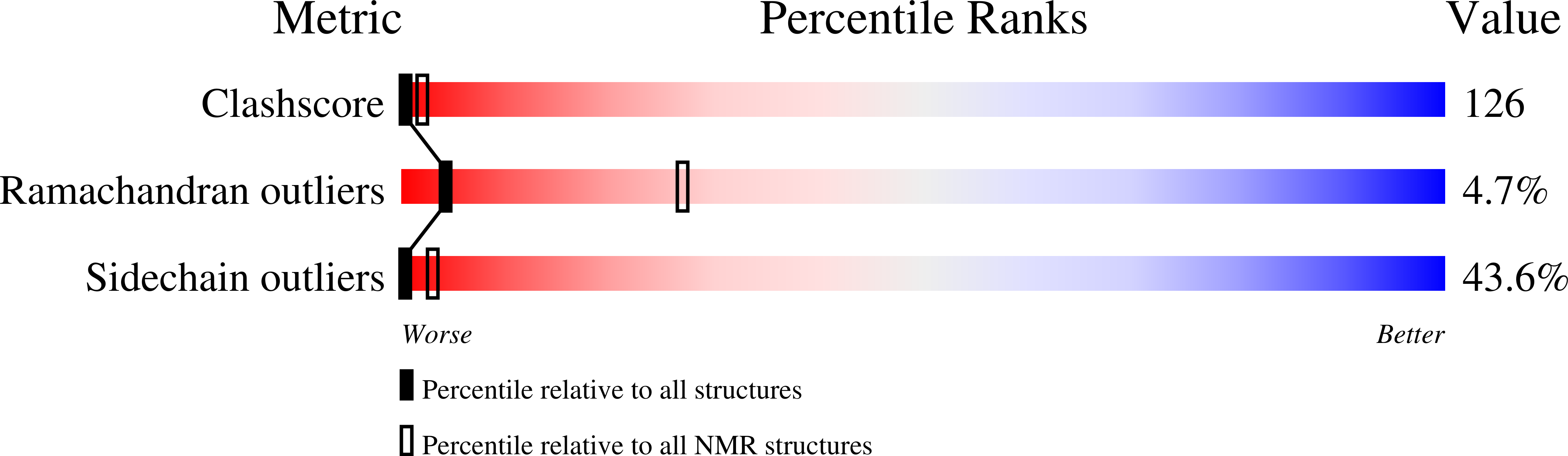Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids.
Liu, Y.J., Cheng, C.S., Lai, S.M., Hsu, M.P., Chen, C.S., Lyu, P.C.(2006) Proteins 63: 777-786
- PubMed: 16544327
- DOI: https://doi.org/10.1002/prot.20962
- Primary Citation of Related Structures:
1TI5 - PubMed Abstract:
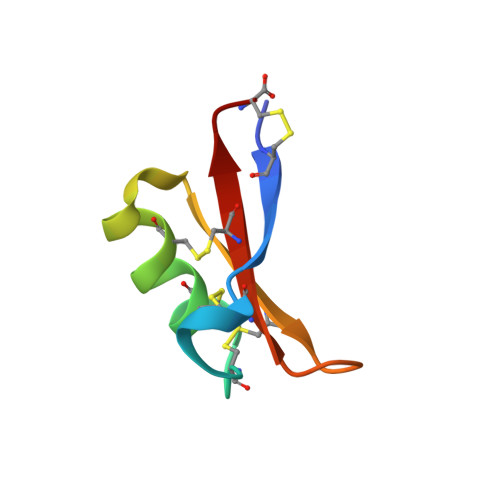
Vigna radiata plant defensin 1 (VrD1) is the first reported plant defensin exhibiting insecticidal activity. We report herein the nuclear magnetic resonance solution structure of VrD1 and the implication on its insecticidal activity. The root-mean-square deviation values are 0.51 +/- 0.35 and 1.23 +/- 0.29 A for backbone and all heavy atoms, respectively. The VrD1 structure comprises a triple-stranded antiparallel beta-sheet, an alpha-helix, and a 3(10) helix stabilized by four disulfide bonds, forming a typical cysteine-stabilized alphabeta motif. Among plant defensins of known structure, VrD1 is the first to contain a 3(10) helix. Glu26 is highly conserved among defensins; VrD1 contains an arginine at this position, which may induce a shift in the orientation of Trp10, thereby promoting the formation of this 3(10) helix. Moreover, VrD1 inhibits Tenebrio molitor alpha-amylase. Alpha-amylase has an essential role in the digestion of plant starch in the insect gut, and expression of the common bean alpha-amylase inhibitor 1 in transgenic pea imparts complete resistance against bruchids. These results imply that VrD1 insecticidal activity has its basis in the inhibition of a polysaccharide hydrolase. Sequence and structural comparisons between two groups of plant defensins having different specificity toward insect alpha-amylase reveal that the loop between beta2 and beta3 is the probable binding site for the alpha-amylase. Computational docking experiments were used to study VrD1-alpha-amylase interactions, and these results provide information that may be used to improve the insecticidal activity of VrD1.
Organizational Affiliation:
Department of Life Sciences, National Tsing Hua University, Hsinchu, Taiwan.