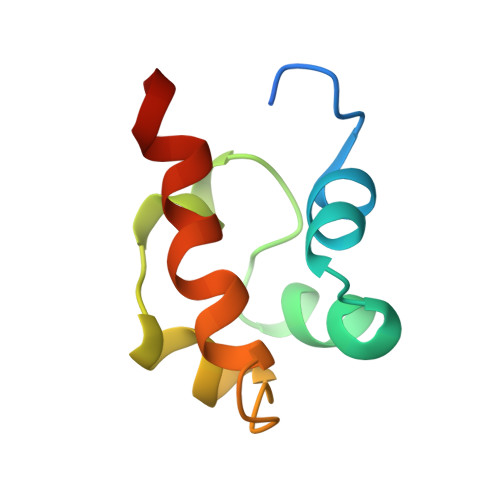
Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites.
Smalla, M., Schmieder, P., Kelly, M., Ter Laak, A., Krause, G., Ball, L., Wahl, M., Bork, P., Oschkinat, H.(1999) Protein Sci 8: 1954-1961
- PubMed: 10548040
- DOI: https://doi.org/10.1110/ps.8.10.1954
- Primary Citation of Related Structures:
1SGG - PubMed Abstract:
The sterile alpha motif (SAM) is a protein interaction domain of around 70 amino acids present predominantly in the N- and C-termini of more than 60 diverse proteins that participate in signal transduction and transcriptional repression. SAM domains have been shown to homo- and hetero-oligomerize and to mediate specific protein-protein interactions. A highly conserved subclass of SAM domains is present at the intracellular C-terminus of more than 40 Eph receptor tyrosine kinases that are involved in the control of axonal pathfinding upon ephrin-induced oligomerization and activation in the event of cell-cell contacts. These SAM domains appear to participate in downstream signaling events via interactions with cytosolic proteins. We determined the solution structure of the EphB2 receptor SAM domain and studied its association behavior. The structure consists of five helices forming a compact structure without binding pockets or exposed conserved aromatic residues. Concentration-dependent chemical shift changes of NMR signals reveal two distinct well-separated areas on the domains' surface sensitive to the formation of homotypic oligomers in solution. These findings are supported by analytical ultracentrifugation studies. The conserved Tyr932, which was reported to be essential for the interaction with SH2 domains after phosphorylation, is buried in the hydrophobic core of the structure. The weak capability of the isolated EphB2 receptor SAM domain to form oligomers is supposed to be relevant in vivo when the driving force of ligand binding induces receptor oligomerization. A formation of SAM tetramers is thought to provide an appropriate contact area for the binding of a low-molecular-weight phosphotyrosine phosphatase and to initiate further downstream responses.
Organizational Affiliation:
Forschungsinstitut für Molekulare Pharmakologie, Berlin-Friedrichsfelde, Germany.