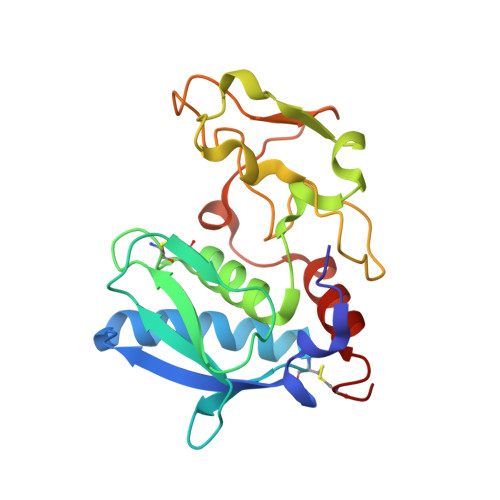
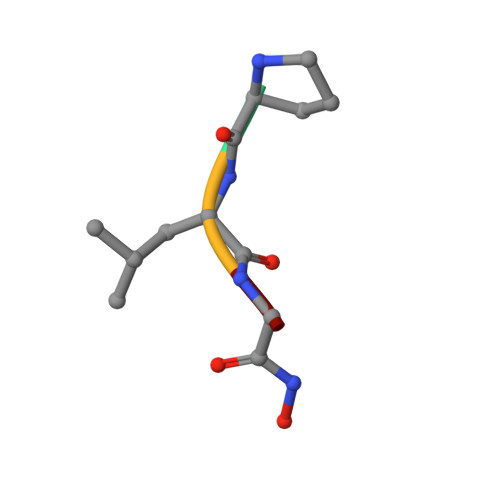
Structure of Astacin with a Transition-State Analogue Inhibitor
Grams, F., Dive, V., Yiotakis, A., Yiallouros, I., Vassiliou, S., Zwilling, R., Bode, W., Stocker, W.(1996) Nat Struct Biol 3: 671
- PubMed: 8756323
- DOI: https://doi.org/10.1038/nsb0896-671
- Primary Citation of Related Structures:
1QJI, 1QJJ