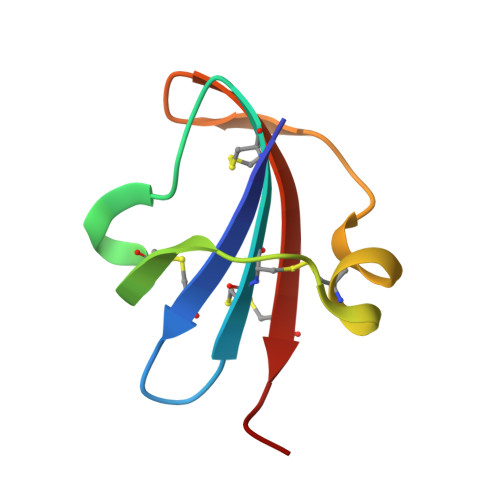
Structure of a novel leech carboxypeptidase inhibitor determined free in solution and in complex with human carboxypeptidase A2.
Reverter, D., Fernandez-Catalan, C., Baumgartner, R., Pfander, R., Huber, R., Bode, W., Vendrell, J., Holak, T.A., Aviles, F.X.(2000) Nat Struct Biol 7: 322-328
- PubMed: 10742178
- DOI: https://doi.org/10.1038/74092
- Primary Citation of Related Structures:
1DTD, 1DTV - PubMed Abstract:
Leech carboxypeptidase inhibitor (LCI) is a novel protein inhibitor present in the medicinal leech Hirudo medicinalis. The structures of LCI free and bound to carboxypeptidase A2 (CPA2)have been determined by NMR and X-ray crystallography, respectively. The LCI structure defines a new protein motif that comprises a five-stranded antiparallel beta-sheet and one short alpha-helix. This structure is preserved in the complex with human CPA2 in the X-ray structure, where the contact regions between the inhibitor and the protease are defined. The C-terminal tail of LCI becomes rigid upon binding the protease as shown in the NMR relaxation studies, and it interacts with the carboxypeptidase in a substrate-like manner. The homology between the C-terminal tails of LCI and the potato carboxypeptidase inhibitor represents a striking example of convergent evolution dictated by the target protease. These new structures are of biotechnological interest since they could elucidate the control mechanism of metallo-carboxypeptidases and could be used as lead compounds for the search of fibrinolytic drugs.
Organizational Affiliation:
Department de Bioquímica i Biologia Molecular, Unitat de Ciències and Institut de Biologia Fonamental, Universitat Autonoma de Barcelona, 08193 Bellaterra, Barcelona, Spain.